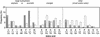Differential proteasomal processing of hydrophobic and hydrophilic protein regions: contribution to cytotoxic T lymphocyte epitope clustering in HIV-1-Nef - PubMed (original) (raw)
Differential proteasomal processing of hydrophobic and hydrophilic protein regions: contribution to cytotoxic T lymphocyte epitope clustering in HIV-1-Nef
Maria Lucchiari-Hartz et al. Proc Natl Acad Sci U S A. 2003.
Abstract
HIV proteins contain a multitude of naturally processed cytotoxic T lymphocyte (CTL) epitopes that concentrate in clusters. The molecular basis of epitope clustering is of interest for understanding HIV immunogenicity and for vaccine design. We show that the CTL epitope clusters of HIV proteins predominantly coincide with hydrophobic regions, whereas the noncluster regions are predominantly hydrophilic. Analysis of the proteasomal degradation products of full-length HIV-Nef revealed a differential sensitivity of cluster and noncluster regions to proteasomal processing. Compared with the epitope-scarce noncluster regions, cluster regions are digested by proteasomes more intensively and with greater preference for hydrophobic P1 residues, resulting in substantially greater numbers of fragments with the sizes and COOH termini typical of epitopes and their precursors. Indeed, many of these fragments correspond to endogenously processed Nef epitopes and/or their potential precursors. The results suggest that differential proteasomal processing contributes importantly to the clustering of CTL epitopes in hydrophobic regions.
Figures
Fig. 1.
Map of the HIV-1-Nef protein showing CTL epitope peptides and proteasomal cleavage products. CTL epitope peptides are shown above the sequence. Black lines are those recognized by CTLs of HIV-infected subjects as recorded in the HIV Molecular Immunology Database. Red lines are the naturally processed peptides acid-eluted from Nef-transfected cells (see also Table 1). The 20S proteasomal cleavage products of recombinant HIV-1-Nef (LAI) protein are indicated as lines below the sequence. Fragments corresponding to acid-eluted HLA class I ligands are highlighted in red; fragments corresponding to their potential precursor peptides are highlighted in green. *, Epitopes identified by immunization of HLA-A2 transgenic mice with HIV-1-Nef (HXB3), with the sequences ALTSSNTA and MTYKAALDL instead of AITSSNTA and MTYKAAVDL (LAI); **, a 20-aa sequence (Nef161–180) where a CTL epitope of unknown HLA restriction was located; ***, subdominant epitope with the sequence DPEKEVLQWK (LAI sequence, DPEREVLEWR).
Fig. 2.
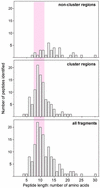
Length distribution of 20S proteasomal cleavage fragments from HIV-1-Nef protein. Recombinant HIV-1-Nef (LAI) protein was digested with purified 20S proteasomes, and the cleavage fragments were determined by MS (see Fig. 1). The preferred size range of MHC class I peptide ligands is highlighted in pink.
Fig. 3.
Frequencies of amino acids at the COOH and NH2 termini of the 143 Nef proteasomal cleavage fragments shown in Fig. 1. The COOH-terminal amino acids are shown as gray bars, and the NH2-terminal amino acids are shown as white bars. For the COOH-terminal amino acids, the proportion of peptides from the CTL epitope clusters is indicated by the hatched parts of the bars. The frequencies (%) of individual amino acids in the Nef protein are given below the x axis.
Fig. 4.
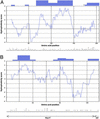
Hydrophobicity/hydrophilicity plots and conservation of the primary sequence of HIV-1-Nef (A) and HIV-1-Gag p17 (B). Hydrophobicity/hydrophilicity plots were generated with the algorithm HPLC/retention pH 7.4; similar plots were obtained with the algorithm of Abraham and Leo (ExPASy Molecular Biology Server,
www.expasy.org/cgi-bin/protscale.pl
). Plots created with window sizes between 15 and 21 looked almost identical. CTL epitope clusters and single CTL epitopes are represented by boxes at the top. For Nef-CTL epitopes, see Fig. 1; for Gag CTL epitopes, see the HIV Molecular Immunology Database. The variability plots (below the hydrophobicity plots) show the number of different amino acids (dots) at each position of the concensus sequences of subtypes A–D. Sequence alignment was performed by using
align
(
www2.igh.cnrs.fr/bin/alignguess.cgi
). *, a 20-aa sequence (Nef161–180) where an HLA class I-restricted epitope (of unknown type restriction) was mapped.
Similar articles
- Cytotoxic T lymphocyte epitopes of HIV-1 Nef: Generation of multiple definitive major histocompatibility complex class I ligands by proteasomes.
Lucchiari-Hartz M, van Endert PM, Lauvau G, Maier R, Meyerhans A, Mann D, Eichmann K, Niedermann G. Lucchiari-Hartz M, et al. J Exp Med. 2000 Jan 17;191(2):239-52. doi: 10.1084/jem.191.2.239. J Exp Med. 2000. PMID: 10637269 Free PMC article. - Characteristics of HIV-1 Nef regions containing multiple CD8+ T cell epitopes: wealth of HLA-binding motifs and sensitivity to proteasome degradation.
Choppin J, Cohen W, Bianco A, Briand JP, Connan F, Dalod M, Guillet JG. Choppin J, et al. J Immunol. 2001 May 15;166(10):6164-9. doi: 10.4049/jimmunol.166.10.6164. J Immunol. 2001. PMID: 11342637 - HLA supertypes contribute in HIV type 1 cytotoxic T lymphocyte epitope clustering in Nef and Gag proteins.
Chakraborty S, Rahman T, Chakravorty R, Kuchta A, Rabby A, Sahiuzzaman M. Chakraborty S, et al. AIDS Res Hum Retroviruses. 2013 Feb;29(2):270-8. doi: 10.1089/AID.2012.0160. Epub 2013 Jan 8. AIDS Res Hum Retroviruses. 2013. PMID: 23061377 - The specificity of proteasomes: impact on MHC class I processing and presentation of antigens.
Niedermann G, Geier E, Lucchiari-Hartz M, Hitziger N, Ramsperger A, Eichmann K. Niedermann G, et al. Immunol Rev. 1999 Dec;172:29-48. doi: 10.1111/j.1600-065x.1999.tb01354.x. Immunol Rev. 1999. PMID: 10631935 Review. - [The HIV nef and the Kaposi-sarcoma-associated virus K3/K5 proteins: "parasites"of the endocytosis pathway].
Bénichou S, Benmerah A. Bénichou S, et al. Med Sci (Paris). 2003 Jan;19(1):100-6. doi: 10.1051/medsci/2003191100. Med Sci (Paris). 2003. PMID: 12836198 Review. French.
Cited by
- Development of multivalent vaccine targeting M segment of Crimean Congo Hemorrhagic Fever Virus (CCHFV) using immunoinformatic approaches.
Sana M, Javed A, Babar Jamal S, Junaid M, Faheem M. Sana M, et al. Saudi J Biol Sci. 2022 Apr;29(4):2372-2388. doi: 10.1016/j.sjbs.2021.12.004. Epub 2021 Dec 10. Saudi J Biol Sci. 2022. PMID: 35531180 Free PMC article. - Cell type-specific proteasomal processing of HIV-1 Gag-p24 results in an altered epitope repertoire.
Steers NJ, Currier JR, Kijak GH, di Targiani RC, Saxena A, Marovich MA, Kim JH, Michael NL, Alving CR, Rao M. Steers NJ, et al. J Virol. 2011 Feb;85(4):1541-53. doi: 10.1128/JVI.01790-10. Epub 2010 Nov 24. J Virol. 2011. PMID: 21106750 Free PMC article. - Characterization of the Protective HIV-1 CTL Epitopes and the Corresponding HLA Class I Alleles: A Step towards Designing CTL Based HIV-1 Vaccine.
Chakraborty S, Rahman T, Chakravorty R. Chakraborty S, et al. Adv Virol. 2014;2014:321974. doi: 10.1155/2014/321974. Epub 2014 Mar 18. Adv Virol. 2014. PMID: 24744786 Free PMC article. Review. - Cross-sectional analysis of CD8 T cell immunity to human herpesvirus 6B.
Martin LK, Hollaus A, Stahuber A, Hübener C, Fraccaroli A, Tischer J, Schub A, Moosmann A. Martin LK, et al. PLoS Pathog. 2018 Apr 26;14(4):e1006991. doi: 10.1371/journal.ppat.1006991. eCollection 2018 Apr. PLoS Pathog. 2018. PMID: 29698478 Free PMC article. - The distribution of CTL epitopes in HIV-1 appears to be random, and similar to that of other proteomes.
Schmid BV, Keşmir C, de Boer RJ. Schmid BV, et al. BMC Evol Biol. 2009 Aug 4;9:184. doi: 10.1186/1471-2148-9-184. BMC Evol Biol. 2009. PMID: 19653887 Free PMC article.
References
- Rowland-Jones, S., Tan, R. & McMichael, A. (1997) Adv. Immunol. 65, 277-346. - PubMed
- Zhang, C., Cornette, J. L., Berzofsky, J. A. & DeLisi, C. (1997) Vaccine 15, 1291-1302. - PubMed
- Kuiken, C. L., Foley, B. T., Guzman, E. & Korber, B. T. (1999) in Molecular Evolution of HIV, ed. Crandall, K. (Johns Hopkins Univ. Press, Baltimore), pp. 432-468.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources