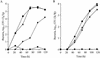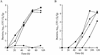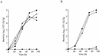Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila - PubMed (original) (raw)
Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila
Tal Zusman et al. Infect Immun. 2003 Jul.
Abstract
Coxiella burnetii, the etiological agent of Q fever, is an obligate intracellular pathogen, whereas Legionella pneumophila, the causative agent of Legionnaires' disease, is a facultative intracellular pathogen. During infection of humans both of these pathogens multiply in alveolar macrophages inside a closed phagosome. L. pneumophila intracellular multiplication was shown to be dependent on the icm/dot system, which probably encodes a type IV-related translocation apparatus. Recently, genes homologous to all of the L. pneumophila icm/dot genes (besides icmR) were found in C. burnetii. To explore the similarities and differences between the icm/dot pathogenesis systems of these two pathogens, interspecies complementation analysis was performed. Nine C. burnetii icm homologous genes (icmT, icmS, icmQ, icmP, icmO, icmJ, icmB, icmW, and icmX) were cloned under regulation of the corresponding L. pneumophila icm genes and examined for the ability to complement L. pneumophila mutants with mutations in these genes. The C. burnetii icmS and icmW homologous genes were found to complement the corresponding L. pneumophila icm mutants to wild-type levels of intracellular growth in both HL-60-derived human macrophages and Acanthamoeba castellanii. In addition, the C. burnetii icmT homologous gene was found to completely complement an L. pneumophila insertion mutant for intracellular growth in HL-60-derived human macrophages, but it only partially complemented the same mutant for intracellular growth in A. castellanii. Moreover, as previously shown for L. pneumophila, the proteins encoded by the C. burnetii icmS and icmW homologous genes were found to interact with one another, and interspecies protein interaction was observed as well. Our results strongly indicate that the Icm/Dot pathogenesis systems of C. burnetii and L. pneumophila have common features.
Figures
FIG. 1.
Complementation analysis with the C. burnetii icmT homologous gene. Intracellular growth experiments in the protozoan host A. castellanii (A) and HL-60-derived human macrophages (B) were performed as described in Materials and Methods. Wild-type L. pneumophila JR-32 (⧫) and the icmT mutant (GS3011) containing the vector pMMB207αb-Km-14 (▪), the L. pneumophila icmTS operon (pGS-Lc-37-14) (▴), and the C. burnetii icmTS homologous genes under regulation of the L. pneumophila icmTS regulatory region (pZT-lpncox-TS) (•) were used. The experiments were performed at least three times, and similar results were obtained each time.
FIG. 2.
C. burnetii icmS homologous gene complements an L. pneumophila icmS mutant for intracellular growth in two hosts. Intracellular growth experiments in the protozoan host A. castellanii (A) and in HL-60-derived human macrophages (B) were performed as described in Materials and Methods. Wild-type L. pneumophila JR-32 (⧫) the icmS mutant (GS3001) containing the vector pMMB207αb-Km-14 (▪), the L. pneumophila icmTS operon (pGS-Lc-37-14) (▴), and the C. burnetii icmTS homologous genes under regulation of the L. pneumophila icmTS regulatory region (pZT-lpncox-TS) (•), and the 25D mutant (□) were used. The experiments were performed three times, and similar results were obtained each time.
FIG. 3.
C. burnetii icmW homologous gene complements an L. pneumophila icmW mutant for intracellular growth. Intracellular growth experiments in the protozoan host A. castellanii were performed as described in Materials and Methods. (A) Wild-type L. pneumophila JR-32 (⧫) and the icmW mutant (GY141) containing the vector pMMB207αb-Km-14 (▪), the L. pneumophila icmW gene (pZT-W-01) (▵), the L. pneumophila icmWX operon (pZT-WX-01) (▴), and the C. burnetii icmWX homologous genes under regulation of the L. pneumophila icmWX regulatory region (pZT-lpncox-WX) (•) were used. (B) icmX mutant LELA3993 containing the same plasmids (without pZT-W-01) was used. The experiments were performed three times, and similar results were obtained each time.
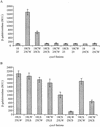
FIG. 4.
Two-hybrid analysis of the interaction between the C. burnetii and L. pneumophila IcmW and IcmS proteins. An E. coli cyaA deletion strain containing various pairs of plasmids was examined for β-galactosidase activity, as described in Materials and Methods. (A) Analysis of the C. burnetii IcmS-IcmW homologous protein interaction; (B) analysis of the L. pneumophila IcmS-IcmW protein interaction and interspecies protein interaction. The plasmids used were the vector pT18 (18) the vector pT25 (25), the T18 fragment fused to the C. burnetii icmS gene (18CS), the T18 fragment fused to the C. burnetii icmW gene (18CW), the T25 fragment fused to the C. burnetii icmS gene (25CS), and the T25 fragment fused to the C. burnetii icmW gene (25CW). Fusions with the L. pneumophila icmS and icmW genes are indicated by L instead of C before the letter that indicates the gene. The data are expressed in Miller units (M.U.) and are the averages ± standard deviations (error bars) of at least three different experiments.
Similar articles
- Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system.
Zamboni DS, McGrath S, Rabinovitch M, Roy CR. Zamboni DS, et al. Mol Microbiol. 2003 Aug;49(4):965-76. doi: 10.1046/j.1365-2958.2003.03626.x. Mol Microbiol. 2003. PMID: 12890021 - The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii.
Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. Zusman T, et al. Mol Microbiol. 2007 Mar;63(5):1508-23. doi: 10.1111/j.1365-2958.2007.05604.x. Mol Microbiol. 2007. PMID: 17302824 - The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii.
Segal G, Feldman M, Zusman T. Segal G, et al. FEMS Microbiol Rev. 2005 Jan;29(1):65-81. doi: 10.1016/j.femsre.2004.07.001. FEMS Microbiol Rev. 2005. PMID: 15652976 Review. - Legionella and Coxiella effectors: strength in diversity and activity.
Qiu J, Luo ZQ. Qiu J, et al. Nat Rev Microbiol. 2017 Oct;15(10):591-605. doi: 10.1038/nrmicro.2017.67. Epub 2017 Jul 17. Nat Rev Microbiol. 2017. PMID: 28713154 Review.
Cited by
- Vaccinomics to Design a Multiepitope Vaccine against Legionella pneumophila.
Umar A, Liaquat S, Fatima I, Rehman A, Rasool D, Alshammari A, Alharbi M, Rajoka MSR, Khurshid M, Ashfaq UA, Haque A. Umar A, et al. Biomed Res Int. 2022 Sep 17;2022:4975721. doi: 10.1155/2022/4975721. eCollection 2022. Biomed Res Int. 2022. PMID: 36164443 Free PMC article. - Identification of two Legionella pneumophila effectors that manipulate host phospholipids biosynthesis.
Viner R, Chetrit D, Ehrlich M, Segal G. Viner R, et al. PLoS Pathog. 2012;8(11):e1002988. doi: 10.1371/journal.ppat.1002988. Epub 2012 Nov 1. PLoS Pathog. 2012. PMID: 23133385 Free PMC article. - Comparative genome analysis of "Candidatus Phytoplasma australiense" (subgroup tuf-Australia I; rp-A) and "Ca. Phytoplasma asteris" Strains OY-M and AY-WB.
Tran-Nguyen LT, Kube M, Schneider B, Reinhardt R, Gibb KS. Tran-Nguyen LT, et al. J Bacteriol. 2008 Jun;190(11):3979-91. doi: 10.1128/JB.01301-07. Epub 2008 Mar 21. J Bacteriol. 2008. PMID: 18359806 Free PMC article. - Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system.
Feldman M, Zusman T, Hagag S, Segal G. Feldman M, et al. Proc Natl Acad Sci U S A. 2005 Aug 23;102(34):12206-11. doi: 10.1073/pnas.0501850102. Epub 2005 Aug 9. Proc Natl Acad Sci U S A. 2005. PMID: 16091472 Free PMC article. - The Coxiella burnetii effector EmcB is a deubiquitinase that inhibits RIG-I signaling.
Duncan-Lowey J, Crabill E, Jarret A, Reed SCO, Roy CR. Duncan-Lowey J, et al. Proc Natl Acad Sci U S A. 2023 Mar 14;120(11):e2217602120. doi: 10.1073/pnas.2217602120. Epub 2023 Mar 9. Proc Natl Acad Sci U S A. 2023. PMID: 36893270 Free PMC article.
References
- Berger, K. H., J. J. Merriam, and R. R. Isberg. 1994. Altered intracellular targeting properties associated with mutations in the Legionella dotA gene. Mol. Microbiol. 14:809-822. - PubMed
- Brand, B. C., A. B. Sadosky, and H. A. Shuman. 1994. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol. Microbiol. 14:797-808. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources