Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae - PubMed (original) (raw)
Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae
Luz Balsalobre et al. Antimicrob Agents Chemother. 2003 Jul.
Abstract
A total of 46 ciprofloxacin-resistant (Cip(r)) Streptococcus pneumoniae strains were isolated from 1991 to 2001 at the Hospital of Bellvitge. Five of these strains showed unexpectedly high rates of nucleotide variations in the quinolone resistance-determining regions (QRDRs) of their parC, parE, and gyrA genes. The nucleotide sequence of the full-length parC, parE, and gyrA genes of one of these isolates revealed a mosaic structure compatible with an interspecific recombination origin. Southern blot analysis and nucleotide sequence determinations showed the presence of an ant-like gene in the intergenic parE-parC regions of the S. pneumoniae Cip(r) isolates with high rates of variations in their parE and parC QRDRs. The ant-like gene was absent from typical S. pneumoniae strains, whereas it was present in the intergenic parE-parC regions of the viridans group streptococci (Streptococcus mitis and Streptococcus oralis). These results suggest that the viridans group streptococci are acting as donors in the horizontal transfer of fluoroquinolone resistance genes to S. pneumoniae.
Figures
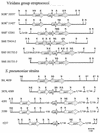
FIG. 1.
Nucleotide sequence variations in the ParC, ParE, and GyrA QRDRs. The nucleotides present at each polymorphic site are shown for strain R6, but for the other strains, only the nucleotides that differ from those in R6 are shown. Changes yielding amino acid substitutions, including those involved in fluoroquinolone resistance, are shown in boldface. Codon numbers are indicated vertically above the sequences. Positions 1, 2, and 3 in the fourth row refer to the first, second, and third nucleotides in the codon, respectively. Strains whose sequences do not differ from that of R6 more than 1% are shaded in gray. SPN, S. pneumoniae; SOR, S. oralis; SMI, S. mitis.
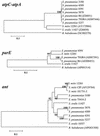
FIG. 2.
UPGMA trees of concatenated atpC and atpA genes, full-length parE genes, and ant genes. Phylogenetic and molecular evolutionary analyses were conducted with the MEGA program (version 2.1) by the UPGMA method. Only bootstrap confidence intervals exceeding 90% are shown. The GenBank accession numbers for each nucleotide sequence are shown in parentheses.
FIG. 3.
Mosaic structures of the gyrA, parC, and parE genes of the indicated strains of S. pneumoniae and VS. The locations of the QRDRs are indicated at the tops of the gyrA and parC sequences. The positions of the active Tyr residues (Y120 in GyrA and Y118 in ParC) that bind to DNA and the Ser residues that are changed in strain 4391 (S81 in GyrA and S79 in ParC) and that are involved in resistance are marked. Blocks showing the percent sequence divergence from the corresponding regions of S. pneumoniae R6 are indicated. White box, region of the sequence that differed by ≤1.5%; gray boxes, regions that differed by more than 1.5% but less than 9%; black boxes, regions that differed by >9%. The strains used were S. pneumoniae (SPN) R6 (GenBank accession number AE008451), S. pneumoniae TIGR4 (GenBank accession number AE007391), S. pneumoniae 4391, S. mitis (SMI) NCTC 12261, and S. oralis (SOR) NCTC 11427.
FIG. 4.
Restriction map of the _parE_-parC region of S. pneumoniae strains and strains of VS and its genetic organization as deduced from Southern blotting experiments and nucleotide sequence analyses. E, _Eco_RV; N, _Nco_I. The parE and parC genes with mosaic structures are indicated by with striped arrows. SOR, S. oralis; SMI, S. mitis.
FIG. 5.
Southern blot hybridization of S. pneumoniae strains and strains of VS with an _ant_-specific probe. Chromosomal DNAs from the indicated strains were cleaved with _Eco_RV-_Nco_I, and the fragments were separated in 0.8% agarose gels. The gels were blotted, and the blot was probed with a biotinylated probe derived from S. pneumoniae 3870 containing positions 26 to 290 of the ant gene. The numbers on the bottom right are molecular sizes (in kilobases).
FIG. 5.
Southern blot hybridization of S. pneumoniae strains and strains of VS with an _ant_-specific probe. Chromosomal DNAs from the indicated strains were cleaved with _Eco_RV-_Nco_I, and the fragments were separated in 0.8% agarose gels. The gels were blotted, and the blot was probed with a biotinylated probe derived from S. pneumoniae 3870 containing positions 26 to 290 of the ant gene. The numbers on the bottom right are molecular sizes (in kilobases).
Similar articles
- Fitness of Streptococcus pneumoniae fluoroquinolone-resistant strains with topoisomerase IV recombinant genes.
Balsalobre L, de la Campa AG. Balsalobre L, et al. Antimicrob Agents Chemother. 2008 Mar;52(3):822-30. doi: 10.1128/AAC.00731-07. Epub 2007 Dec 26. Antimicrob Agents Chemother. 2008. PMID: 18160515 Free PMC article. - Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci.
González I, Georgiou M, Alcaide F, Balas D, Liñares J, de la Campa AG. González I, et al. Antimicrob Agents Chemother. 1998 Nov;42(11):2792-8. doi: 10.1128/AAC.42.11.2792. Antimicrob Agents Chemother. 1998. PMID: 9797205 Free PMC article. - Fluoroquinolone resistance in atypical pneumococci and oral streptococci: evidence of horizontal gene transfer of fluoroquinolone resistance determinants from Streptococcus pneumoniae.
Ip M, Chau SS, Chi F, Tang J, Chan PK. Ip M, et al. Antimicrob Agents Chemother. 2007 Aug;51(8):2690-700. doi: 10.1128/AAC.00258-07. Epub 2007 Jun 4. Antimicrob Agents Chemother. 2007. PMID: 17548487 Free PMC article. - Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance.
Pan XS, Fisher LM. Pan XS, et al. J Bacteriol. 1996 Jul;178(14):4060-9. doi: 10.1128/jb.178.14.4060-4069.1996. J Bacteriol. 1996. PMID: 8763932 Free PMC article.
Cited by
- Impact of fluoroquinolone resistance mutations on gonococcal fitness and in vivo selection for compensatory mutations.
Kunz AN, Begum AA, Wu H, D'Ambrozio JA, Robinson JM, Shafer WM, Bash MC, Jerse AE. Kunz AN, et al. J Infect Dis. 2012 Jun 15;205(12):1821-9. doi: 10.1093/infdis/jis277. Epub 2012 Apr 5. J Infect Dis. 2012. PMID: 22492860 Free PMC article. - Characterization of recombinant fluoroquinolone-resistant pneumococcus-like isolates.
Balsalobre L, Ortega M, de la Campa AG. Balsalobre L, et al. Antimicrob Agents Chemother. 2013 Jan;57(1):254-60. doi: 10.1128/AAC.01357-12. Epub 2012 Oct 31. Antimicrob Agents Chemother. 2013. PMID: 23114769 Free PMC article. - Independent behavior of commensal flora for carriage of fluoroquinolone-resistant bacteria in patients at admission.
de Lastours V, Chau F, Tubach F, Pasquet B, Ruppé E, Fantin B. de Lastours V, et al. Antimicrob Agents Chemother. 2010 Dec;54(12):5193-200. doi: 10.1128/AAC.00823-10. Epub 2010 Sep 27. Antimicrob Agents Chemother. 2010. PMID: 20876373 Free PMC article. - Identifying mutator phenotypes among fluoroquinolone-resistant strains of Streptococcus pneumoniae using fluctuation analysis.
Gould CV, Sniegowski PD, Shchepetov M, Metlay JP, Weiser JN. Gould CV, et al. Antimicrob Agents Chemother. 2007 Sep;51(9):3225-9. doi: 10.1128/AAC.00336-07. Epub 2007 Jul 30. Antimicrob Agents Chemother. 2007. PMID: 17664329 Free PMC article. - New species genetic approach to identify strains of mitis group streptococci that are donors of rifampin resistance to Streptococcus pneumoniae.
Ferrándiz MJ, Ardanuy C, Liñares J, Balsalobre L, García MT, de la Campa AG. Ferrándiz MJ, et al. Antimicrob Agents Chemother. 2011 Jan;55(1):368-72. doi: 10.1128/AAC.00856-10. Epub 2010 Nov 1. Antimicrob Agents Chemother. 2011. PMID: 21041504 Free PMC article.
References
- Alcaide, F., J. Carratala, J. Liñares, F. Gudiol, and R. Martín. 1996. In vitro activities of eight macrolide antibiotics and RP-59500 (quinupristin-dalfopristin) against viridans group streptococci isolated from blood of neutropenic cancer patients. Antimicrob. Agents Chemother. 40:2117-2120. - PMC - PubMed
- Alcaide, F., J. Liñares, R. Pallarés, J. Carratala, M. A. Benítez, F. Gudiol, and R. Martín. 1995. In vitro activity of 22 β-lactam antibiotics against penicillin-resistant and penicillin-susceptible viridans group streptococci isolated from blood. Antimicrob. Agents Chemother. 39:2243-2247. - PMC - PubMed
- Awada, A. P., P. Van der Auwera, P. Meunier, D. Daneau, and J. Klastersky. 1992. Streptococcal and enterococcal bacteremia in patients with cancer. Clin. Infect. Dis. 15:33-48. - PubMed
- Bartlett, J. G., R. F. Breiman, L. Mandell, and T. M. File. 1998. Community-acquired pneumonia in adults: guidelines for management. Clin. Infect. Dis. 26:811-838. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources