Statistical implications of pooling RNA samples for microarray experiments - PubMed (original) (raw)
Statistical implications of pooling RNA samples for microarray experiments
Xuejun Peng et al. BMC Bioinformatics. 2003.
Abstract
Background: Microarray technology has become a very important tool for studying gene expression profiles under various conditions. Biologists often pool RNA samples extracted from different subjects onto a single microarray chip to help defray the cost of microarray experiments as well as to correct for the technical difficulty in getting sufficient RNA from a single subject. However, the statistical, technical and financial implications of pooling have not been explicitly investigated.
Results: Modeling the resulting gene expression from sample pooling as a mixture of individual responses, we derived expressions for the experimental error and provided both upper and lower bounds for its value in terms of the variability among individuals and the number of RNA samples pooled. Using "virtual" pooling of data from real experiments and computer simulations, we investigated the statistical properties of RNA sample pooling. Our study reveals that pooling biological samples appropriately is statistically valid and efficient for microarray experiments. Furthermore, optimal pooling design(s) can be found to meet statistical requirements while minimizing total cost.
Conclusions: Appropriate RNA pooling can provide equivalent power and improve efficiency and cost-effectiveness for microarray experiments with a modest increase in total number of subjects. Pooling schemes in terms of replicates of subjects and arrays can be compared before experiments are conducted.
Figures
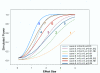
Figure 1
Relationships among type I error rate, sample size, effect size, and power with or without pooling. Note: n is the number of biological replicates per treatment group; c is the number of gene chips per group; a is the type I error rate; EQ means that samples are pooled with equal contribution; NE means samples do not contribute equally when pooled together (weights assigned randomly to each chip: 0.7, 0.2, and 0.1).
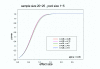
Figure 2
Approximately equivalent power curves under different pooling schemes. Power curves generated for two-sample t tests. Equal pooling assumed. Legend: n is the number of subjects per treatment group; c is the number of arrays per group. The five pooling schemes with different choices of number of subjects and number of arrays have approximately equivalent power curves when type I error rate is controlled at 0.05.
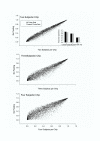
Figure 3
Scatter plots of the P-values with different "virtual" pooling schemes. On Y-axis are the P-values from two-sample t-tests for 8799 genes on the RGU34A gene chip with no pooling (12 subjects, 12 arrays per group). On X axis are the P-values from two-sample t-tests for pool size 2 (12 subjects, 6 arrays per group), pool size 3 (12 subjects, 4 arrays per group), and pool size 4 (12 subjects, 3 arrays per group), respectively.
Similar articles
- Effect of pooling samples on the efficiency of comparative studies using microarrays.
Zhang SD, Gant TW. Zhang SD, et al. Bioinformatics. 2005 Dec 15;21(24):4378-83. doi: 10.1093/bioinformatics/bti717. Epub 2005 Oct 18. Bioinformatics. 2005. PMID: 16234321 - On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments.
Takele Assefa A, Vandesompele J, Thas O. Takele Assefa A, et al. BMC Genomics. 2020 Apr 19;21(1):312. doi: 10.1186/s12864-020-6721-y. BMC Genomics. 2020. PMID: 32306892 Free PMC article. - Effects of pooling mRNA in microarray class comparisons.
Shih JH, Michalowska AM, Dobbin K, Ye Y, Qiu TH, Green JE. Shih JH, et al. Bioinformatics. 2004 Dec 12;20(18):3318-25. doi: 10.1093/bioinformatics/bth391. Epub 2004 Jul 9. Bioinformatics. 2004. PMID: 15247103 - Microarray data analysis: from disarray to consolidation and consensus.
Allison DB, Cui X, Page GP, Sabripour M. Allison DB, et al. Nat Rev Genet. 2006 Jan;7(1):55-65. doi: 10.1038/nrg1749. Nat Rev Genet. 2006. PMID: 16369572 Review. - Toward understanding the informatics and statistical aspects of micro-RNA profiling.
Sarver AL. Sarver AL. J Cardiovasc Transl Res. 2010 Jun;3(3):204-11. doi: 10.1007/s12265-010-9180-z. Epub 2010 May 4. J Cardiovasc Transl Res. 2010. PMID: 20560041 Review.
Cited by
- Dramatic Suppression of Lipogenesis and No Increase in Beta-Oxidation Gene Expression Are among the Key Effects of Bergamot Flavonoids in Fatty Liver Disease.
Parafati M, La Russa D, Lascala A, Crupi F, Riillo C, Fotschki B, Mollace V, Janda E. Parafati M, et al. Antioxidants (Basel). 2024 Jun 25;13(7):766. doi: 10.3390/antiox13070766. Antioxidants (Basel). 2024. PMID: 39061835 Free PMC article. - Differentially Expressed Genes in Response to a Squalene-Supplemented Diet Are Accurate Discriminants of Porcine Non-Alcoholic Steatohepatitis.
Abuobeid R, Herrera-Marcos LV, Arnal C, Bidooki SH, Sánchez-Marco J, Lasheras R, Surra JC, Rodríguez-Yoldi MJ, Martínez-Beamonte R, Osada J. Abuobeid R, et al. Int J Mol Sci. 2023 Aug 8;24(16):12552. doi: 10.3390/ijms241612552. Int J Mol Sci. 2023. PMID: 37628732 Free PMC article. - Modulation of Anthocyanin Biosynthesis-Related Genes during the Ripening of Olea europaea L. cvs Carolea and Tondina Drupes in Relation to Environmental Factors.
Ferrari M, Muto A, Bruno L, Muzzalupo I, Chiappetta A. Ferrari M, et al. Int J Mol Sci. 2023 May 15;24(10):8770. doi: 10.3390/ijms24108770. Int J Mol Sci. 2023. PMID: 37240115 Free PMC article. - Preclinical species gene expression database: Development and meta-analysis.
Krause C, Suwada K, Blomme EAG, Kowalkowski K, Liguori MJ, Mahalingaiah PK, Mittelstadt S, Peterson R, Rendino L, Vo A, Van Vleet TR. Krause C, et al. Front Genet. 2023 Jan 17;13:1078050. doi: 10.3389/fgene.2022.1078050. eCollection 2022. Front Genet. 2023. PMID: 36733943 Free PMC article. - Transcriptional profiling of drug-induced liver injury biomarkers: association of hepatic Srebf1/Pparα signaling and crosstalk of thrombin, alcohol dehydrogenase, MDR and DNA damage regulators.
Ghanim BY, Ahmad M, Abdallah Q, Khaleel A, Qinna NA. Ghanim BY, et al. Mol Cell Biochem. 2023 Sep;478(9):1949-1960. doi: 10.1007/s11010-022-04648-1. Epub 2022 Dec 30. Mol Cell Biochem. 2023. PMID: 36583794
References
- Miller RA, Galecki A, Shmookler-Reis RJ. Interpretation, design, and analysis of gene array expression experiments. J of Gerontol A Biol Sci Med Sci. 2001;56:B52–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 RR016481/RR/NCRR NIH HHS/United States
- 1P20RR16481-01/RR/NCRR NIH HHS/United States
- P01 AG010836/AG/NIA NIH HHS/United States
- R37 AG004542/AG/NIA NIH HHS/United States
- AG-10836/AG/NIA NIH HHS/United States
- AG-04542/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources