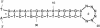Mfold web server for nucleic acid folding and hybridization prediction - PubMed (original) (raw)
Mfold web server for nucleic acid folding and hybridization prediction
Michael Zuker. Nucleic Acids Res. 2003.
Abstract
The abbreviated name, 'mfold web server', describes a number of closely related software applications available on the World Wide Web (WWW) for the prediction of the secondary structure of single stranded nucleic acids. The objective of this web server is to provide easy access to RNA and DNA folding and hybridization software to the scientific community at large. By making use of universally available web GUIs (Graphical User Interfaces), the server circumvents the problem of portability of this software. Detailed output, in the form of structure plots with or without reliability information, single strand frequency plots and 'energy dot plots', are available for the folding of single sequences. A variety of 'bulk' servers give less information, but in a shorter time and for up to hundreds of sequences at once. The portal for the mfold web server is http://www.bioinfo.rpi.edu/applications/mfold. This URL will be referred to as 'MFOLDROOT'.
Figures
Figure 1
This figure reproduces part of the initial results page for an RNA folding. Some input is reproduced in the text area. Various hyperlinks lead to the actual folding results.
Figure 2
Two complementary DNA sequences have been concatenated by linking them with six Ns. The two loops are labeled ‘1’ and ‘2’. Loop ‘1’ is an exterior loop, which is correct, but loop ‘2’ is a hairpin loop instead of an exterior loop.
Similar articles
- UNAFold: software for nucleic acid folding and hybridization.
Markham NR, Zuker M. Markham NR, et al. Methods Mol Biol. 2008;453:3-31. doi: 10.1007/978-1-60327-429-6_1. Methods Mol Biol. 2008. PMID: 18712296 - DINAMelt web server for nucleic acid melting prediction.
Markham NR, Zuker M. Markham NR, et al. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W577-81. doi: 10.1093/nar/gki591. Nucleic Acids Res. 2005. PMID: 15980540 Free PMC article. - Sfold web server for statistical folding and rational design of nucleic acids.
Ding Y, Chan CY, Lawrence CE. Ding Y, et al. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W135-41. doi: 10.1093/nar/gkh449. Nucleic Acids Res. 2004. PMID: 15215366 Free PMC article. - Beyond Mfold: recent advances in RNA bioinformatics.
Reeder J, Höchsmann M, Rehmsmeier M, Voss B, Giegerich R. Reeder J, et al. J Biotechnol. 2006 Jun 25;124(1):41-55. doi: 10.1016/j.jbiotec.2006.01.034. Epub 2006 Mar 10. J Biotechnol. 2006. PMID: 16530285 Free PMC article. Review. - Protein-nucleic acid thermodynamic databases for specific uses.
Mei LC, Hao GF, Yang GF. Mei LC, et al. Trends Biotechnol. 2023 Aug;41(8):990-991. doi: 10.1016/j.tibtech.2023.03.015. Epub 2023 Apr 10. Trends Biotechnol. 2023. PMID: 37045637 Review.
Cited by
- The HIF-1α antisense long non-coding RNA drives a positive feedback loop of HIF-1α mediated transactivation and glycolysis.
Zheng F, Chen J, Zhang X, Wang Z, Chen J, Lin X, Huang H, Fu W, Liang J, Wu W, Li B, Yao H, Hu H, Song E. Zheng F, et al. Nat Commun. 2021 Feb 26;12(1):1341. doi: 10.1038/s41467-021-21535-3. Nat Commun. 2021. PMID: 33637716 Free PMC article. - RNA recognition by the DNA end-binding Ku heterodimer.
Dalby AB, Goodrich KJ, Pfingsten JS, Cech TR. Dalby AB, et al. RNA. 2013 Jun;19(6):841-51. doi: 10.1261/rna.038703.113. Epub 2013 Apr 22. RNA. 2013. PMID: 23610127 Free PMC article. - The Mitogenomes of _Ophiostoma minus and _Ophiostoma piliferum and Comparisons With Other Members of the Ophiostomatales.
Zubaer A, Wai A, Patel N, Perillo J, Hausner G. Zubaer A, et al. Front Microbiol. 2021 Feb 10;12:618649. doi: 10.3389/fmicb.2021.618649. eCollection 2021. Front Microbiol. 2021. PMID: 33643245 Free PMC article. - Downregulation of Nipah virus N mRNA occurs through interaction between its 3' untranslated region and hnRNP D.
Hino K, Sato H, Sugai A, Kato M, Yoneda M, Kai C. Hino K, et al. J Virol. 2013 Jun;87(12):6582-8. doi: 10.1128/JVI.02495-12. Epub 2013 Mar 20. J Virol. 2013. PMID: 23514888 Free PMC article. - Genome Organization and Comparative Evolutionary Mitochondriomics of Brown Planthopper, Nilaparvata lugens Biotype 4 Using Next Generation Sequencing (NGS).
Govindharaj GP, Babu SB, Choudhary JS, Asad M, Chidambaranathan P, Gadratagi BG, Rath PC, Naaz N, Jaremko M, Qureshi KA, Kumar U. Govindharaj GP, et al. Life (Basel). 2022 Aug 23;12(9):1289. doi: 10.3390/life12091289. Life (Basel). 2022. PMID: 36143326 Free PMC article.
References
- Fresco J.R., Alberts,B.M. and Doty,P. (1960) Some molecular details of the secondary structure of ribonucleic acid. Nature, 188, 98–101. - PubMed
- Borer P.N., Dengler,B., Tinoco,I.,Jr and Uhlenbeck,O.C. (1974) Stability of ribonucleic acid double-stranded helices. J. Mol. Biol., 86, 843–853. - PubMed
- Tinoco I., Jr and Uhlenbeck,O.C. (1971) Estimation of secondary structure in ribonucleic acids. Nature, 230, 362–367. - PubMed
- Tinoco I. Jr, Borer,P.N., Dengler,B., Levine,M.D., Uhlenbeck,O.C., Crothers,D.M. and Gralla,J. (1973) Improved estimation of secondary structure in ribonucleic acids. Nature New Biol., 246, 40–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases