Pfold: RNA secondary structure prediction using stochastic context-free grammars - PubMed (original) (raw)
Pfold: RNA secondary structure prediction using stochastic context-free grammars
Bjarne Knudsen et al. Nucleic Acids Res. 2003.
Abstract
RNA secondary structures are important in many biological processes and efficient structure prediction can give vital directions for experimental investigations. Many available programs for RNA secondary structure prediction only use a single sequence at a time. This may be sufficient in some applications, but often it is possible to obtain related RNA sequences with conserved secondary structure. These should be included in structural analyses to give improved results. This work presents a practical way of predicting RNA secondary structure that is especially useful when related sequences can be obtained. The method improves a previous algorithm based on an explicit evolutionary model and a probabilistic model of structures. Predictions can be done on a web server at http://www.daimi.au.dk/\~compbio/pfold.
Figures
Figure 1
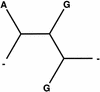
The effect of treating gaps as unknown nucleotides. Only a single column from the alignment is considered with the nucleotides put at the leaves of the phylogenetic tree. The two trees have identical probabilities since leaves with gaps can be removed.
Figure 1
The effect of treating gaps as unknown nucleotides. Only a single column from the alignment is considered with the nucleotides put at the leaves of the phylogenetic tree. The two trees have identical probabilities since leaves with gaps can be removed.
Figure 2
A structure prediction for three hypothetical sequences. In the top alignment, gaps are treated as unknown nucleotides. The structure, shown as parentheses, include pairs between nucleotides and gaps. In the parenthesis notation, corresponding parentheses indicate positions forming base-pairs. In the bottom alignment, the columns with gaps have been left out of the prediction, because <75% of the sequences have nucleotides in these positions.
Figure 2
A structure prediction for three hypothetical sequences. In the top alignment, gaps are treated as unknown nucleotides. The structure, shown as parentheses, include pairs between nucleotides and gaps. In the parenthesis notation, corresponding parentheses indicate positions forming base-pairs. In the bottom alignment, the columns with gaps have been left out of the prediction, because <75% of the sequences have nucleotides in these positions.
Figure 3
In the top alignment, the KH-99 result is given. The bottom alignment shows the structure when all nucleotides have a 1% chance of being any other nucleotide. The result is a longer stem, which includes one non-standard pair.
Figure 3
In the top alignment, the KH-99 result is given. The bottom alignment shows the structure when all nucleotides have a 1% chance of being any other nucleotide. The result is a longer stem, which includes one non-standard pair.
Figure 4
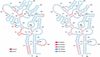
Prediction of the Klebsiella pneumoniae RNase P RNA structure (5) with the KH-99 method based on the four sequence alignment in the work of Knudsen and Hein (12). The left side shows which areas are correctly predicted and the right side shows the reliability of the prediction. Notice the high correlation between the two. Positions 359–366 form a pseudoknot with positions 68–70 and 72–76. Furthermore, positions 84–87 form a pseudoknot with positions 282–285. Since the algorithm described here does not take pseudoknots into account, this explains why these areas are incorrectly predicted while some of them seem reliable.
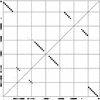
Figure 5
A dot plot made from GCA-tRNA sequences from rat, chicken, mouse and cow (1). The lower left corner represents the beginning of the alignment. Imagining the alignment laid out upwards from here and toward the right, the dots inside the square represent pairing probabilities between positions. The dots outside the square represent probabilities of not pairing. The tRNA structure is clearly visible.
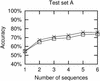
Figure 6
Accuracy as a function of the number of sequences used in the prediction. Crosses are from results using ‘correct’ alignments, while boxes are from ClustalW alignments. Each point represents average results for either all possible combinations of the relevant number of sequences or 50 random combinations, whichever is the lowest number.
Figure 6
Accuracy as a function of the number of sequences used in the prediction. Crosses are from results using ‘correct’ alignments, while boxes are from ClustalW alignments. Each point represents average results for either all possible combinations of the relevant number of sequences or 50 random combinations, whichever is the lowest number.
Figure 6
Accuracy as a function of the number of sequences used in the prediction. Crosses are from results using ‘correct’ alignments, while boxes are from ClustalW alignments. Each point represents average results for either all possible combinations of the relevant number of sequences or 50 random combinations, whichever is the lowest number.
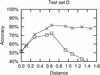
Figure 7
Accuracy as a function of pairwise distance between two sequences being analysed. As in Figure 6, crosses are from results using ‘correct’ alignments, while boxes are from ClustalW alignments. The pairs were grouped according to their Jukes–Cantor distances, in the intervals [0;0.2), [0.2;0.4), [0.4;0.6) etc. The points represent average results for 50 random sequence combinations from a specific range of distances. The _x_-value of a point is the average of the 50 distances.
Similar articles
- Pair stochastic tree adjoining grammars for aligning and predicting pseudoknot RNA structures.
Matsui H, Sato K, Sakakibara Y. Matsui H, et al. Proc IEEE Comput Syst Bioinform Conf. 2004:290-9. Proc IEEE Comput Syst Bioinform Conf. 2004. PMID: 16448022 - Multithreaded comparative RNA secondary structure prediction using stochastic context-free grammars.
Sükösd Z, Knudsen B, Vaerum M, Kjems J, Andersen ES. Sükösd Z, et al. BMC Bioinformatics. 2011 Apr 18;12:103. doi: 10.1186/1471-2105-12-103. BMC Bioinformatics. 2011. PMID: 21501497 Free PMC article. - SCFGs in RNA secondary structure prediction RNA secondary structure prediction: a hands-on approach.
Sükösd Z, Andersen ES, Lyngsø R. Sükösd Z, et al. Methods Mol Biol. 2014;1097:143-62. doi: 10.1007/978-1-62703-709-9_8. Methods Mol Biol. 2014. PMID: 24639159 - Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.
Washietl S, Bernhart SH, Kellis M. Washietl S, et al. Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review. - Introduction to stochastic context free grammars.
Giegerich R. Giegerich R. Methods Mol Biol. 2014;1097:85-106. doi: 10.1007/978-1-62703-709-9_5. Methods Mol Biol. 2014. PMID: 24639156 Review.
Cited by
- Quantifying variances in comparative RNA secondary structure prediction.
Anderson JW, Novák Á, Sükösd Z, Golden M, Arunapuram P, Edvardsson I, Hein J. Anderson JW, et al. BMC Bioinformatics. 2013 May 1;14:149. doi: 10.1186/1471-2105-14-149. BMC Bioinformatics. 2013. PMID: 23634662 Free PMC article. - Detecting and comparing non-coding RNAs in the high-throughput era.
Bussotti G, Notredame C, Enright AJ. Bussotti G, et al. Int J Mol Sci. 2013 Jul 24;14(8):15423-58. doi: 10.3390/ijms140815423. Int J Mol Sci. 2013. PMID: 23887659 Free PMC article. Review. - LinearAlifold: Linear-time consensus structure prediction for RNA alignments.
Malik A, Zhang L, Gautam M, Dai N, Li S, Zhang H, Mathews DH, Huang L. Malik A, et al. J Mol Biol. 2024 Sep 1;436(17):168694. doi: 10.1016/j.jmb.2024.168694. Epub 2024 Jul 4. J Mol Biol. 2024. PMID: 38971557 - Co-transcriptional folding is encoded within RNA genes.
Meyer IM, Miklós I. Meyer IM, et al. BMC Mol Biol. 2004 Aug 6;5:10. doi: 10.1186/1471-2199-5-10. BMC Mol Biol. 2004. PMID: 15298702 Free PMC article. - Computational methods in noncoding RNA research.
Machado-Lima A, del Portillo HA, Durham AM. Machado-Lima A, et al. J Math Biol. 2008 Jan;56(1-2):15-49. doi: 10.1007/s00285-007-0122-6. Epub 2007 Sep 4. J Math Biol. 2008. PMID: 17786447 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources