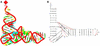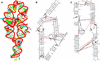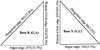Tools for the automatic identification and classification of RNA base pairs - PubMed (original) (raw)
Tools for the automatic identification and classification of RNA base pairs
Huanwang Yang et al. Nucleic Acids Res. 2003.
Abstract
Three programs have been developed to aid in the classification and visualization of RNA structure. BPViewer provides a web interface for displaying three-dimensional (3D) coordinates of individual base pairs or base pair collections. A web server, RNAview, automatically identifies and classifies the types of base pairs that are formed in nucleic acid structures by various combinations of the three edges, Watson-Crick, Hoogsteen and the Sugar edge. RNAView produces two-dimensional (2D) diagrams of secondary and tertiary structure in either Postscript, VRML or RNAML formats. The application RNAMLview can be used to rearrange various parts of the RNAView 2D diagram to generate a standard representation (like the cloverleaf structure of tRNAs) or any layout desired by the user. A 2D diagram can be rapidly reformatted using RNAMLview since all the parts of RNA (like helices and single strands) are dynamically linked while moving the selected parts. With the base pair annotation and the 2D graphic display, RNA motifs are rapidly identified and classified. A survey has been carried out for 41 unique structures selected from the NDB database. The statistics for the occurrence of each edge and of each of the 12 bp families are given for the combinations of the four bases: A, G, U and C. The program also allows for visualization of the base pair interactions by using a symbolic convention previously proposed for base pairs. The web servers for BPViewer and RNAview are available at http://ndbserver.rutgers.edu/services/. The application RNAMLview can also be downloaded from this site. The 2D diagrams produced by RNAview are available for RNA structures in the Nucleic Acid Database (NDB) at http://ndbserver.rutgers.edu/atlas/.
Figures
Figure 1
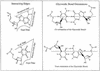
(Left) Identification of edges in RNA bases. (Right) Cis versus trans orientation of glycosidic bonds. The three edges are Waston–Crick, Hoogsteen and Sugar. The trans is formed if the glycosidic bonds of the interacting nucleotides lie on opposite sides of the line. The cis is formed, if the glycosidic bonds of the nucleotides are on the same side of the line. Figure reproduced from (3) with permission from Oxford University Press.
Figure 2
Coordinate frames for each base. For an ideal Watson–Crick pair, the two frames overlap with opposite y and z directions. In this picture, the two frames are separated a little for visualization. The solid circle means the _z_-axis is pointing outside of the paper and the crossed circle means that the _z_-axis is pointing inside of the paper [The picture is taken from Olson (11) with some modification].
Figure 3
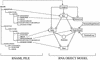
The flowchart of the RNAview program is given at the left and the right drawing gives the scenario of how the RNAview and the RNAMLview programs are integrated.
Figure 4
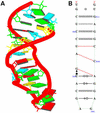
The crystal structure of the sarcin/ricin domain from E.coli 23S rRNA [NDBID UR0007 (16)]. (A) The 3D structure. Each block represents the base of nucleic acid: A, red; C, yellow; G, green; U, cyan. The secondary structure (B) was obtained directly from the RNAview program without further modification. The red single dashed lines mean that the base pairs only involve one H-bond.
Figure 5
Crystal structure of hammerhead ribozyme [NDBID UHX026 (17)]. One biological unit is used for demonstration. (A) The 3D structure. Color code is the same as in Figure 4. The secondary structure (B) was obtained directly from the RNAview program without further modification. The red single dashed lines mean that the base pair only involve one H-bond.
Figure 6
Crystal structure of tRNA [NDBID TR0001 (13)]. (A) The 3D structure. Color code is the same as in Figure 4. The secondary structure (B) was obtained directly from the RNAview program without further modification. The red single dashed lines mean that the base pair involve only one H-bond. The modified bases are given the corresponding lower case letter. The letter P means pseudouracil. The four red letters (a, A, G, g) indicate that the nucleic acids have syn conformations. Modified bases are given as lower case.
Figure 7
The correspondence of the RNAML tree structure with the RNA object model generated and used by RNAMLview.
Figure 8
Schematic illustrating the use of RNAview and RNAMLview. The upper left panel gives the RNAML file generated by RNAview from a PDB file (NDBID TR0001). The upper right panel gives the RNA object model generated by the RNAMLview program. The lower right panel gives the picture of tRNA secondary structure (NDBID TR0001) generated by RNAMLview from the RNA object model. Finally, the lower left panel gives the modified picture created by the RNAMLview.
Figure 9
Crystal structure of mutant P4–P6 domain of Tetrahymena themophila group I intron [NDBID UR0012 (14)]. One monomer (chain A) was selected for demonstration. (A) The 3D structure. Color code is the same as Figure 4a. (B) View obtained directly from the RNAview program without modification. (C) View resulting from modification using RNAMLview. Note that the orientation of (C) matches (A). Red dashed lines means the pair interact by a single H-bond. The red letters indicate that the nucleic acids have syn conformations.
Figure 10
Statistics of the occurrences at each of the three edges for the R and Y bases. They were calculated from the 41 selected structures.
References
- Saenger W. (1983) Principles of Nucleic Acid Structure. Springer-Verlag, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources