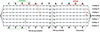FrameD: A flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences - PubMed (original) (raw)
FrameD: A flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences
Thomas Schiex et al. Nucleic Acids Res. 2003.
Abstract
We describe FrameD, a program that predicts coding regions in prokaryotic and matured eukaryotic sequences. Initially targeted at gene prediction in bacterial GC rich genomes, the gene model used in FrameD also allows to predict genes in the presence of frameshifts and partially undetermined sequences which makes it also very suitable for gene prediction and frameshift correction in unfinished sequences such as EST and EST cluster sequences. Like recent eukaryotic gene prediction programs, FrameD also includes the ability to take into account protein similarity information both in its prediction and its graphical output. Its performances are evaluated on different bacterial genomes. The web site (http://genopole.toulouse.inra.fr/bioinfo/FrameD/FD) allows direct prediction, sequence correction and translation and the ability to learn new models for new organisms.
Figures
Figure 1
A simplified view of the directed acyclic graph built for analyzing the sequence CATGAGTACNGA. This view ignores the additional complexity induced by gene overlapping regions and frameshift modeling. The occurrence of a START codon at position 2 to 4 induces a ‘signal’ edge that goes from the non-coding track to the +2 coding track. Similarly, the occurrence of the NGA codon at the end induces a STOP signal edge. Edge weights sources are indicated using dotted arrows.
Figure 2
An example of the graphical output of FrameD. The sequence is on the _x_-axis. The _y_-axis corresponds to possible predictions. From top to bottom: frame 3, 2, 1 coding tracks, intergenic track (IG) and frame −1, −2, −3 coding tracks. In-frame START codons are represented as blue vertical lines. The longer the line, the better a possible RBS. In-frame STOP codons are represented as small red vertical lines (grey if the STOP codon is degenerated). Thin black lines represent the smoothed normalized coding/non coding score. Finally, BlastX hits are represented as magenta blocks. The prediction itself is visible as red blocks and the ‘mean’ prediction as a thin grey line. The thin magenta line represents frameshift expectation. The sequence here has been specifically modified for the example: the ATG start codon of the gene, at position 148 has been replaced by an ANG and 15 nucleotides from position 915 to 929 have been replaced by 14 Ns. Using the ‘low quality sequence’ frameshift penalty, FrameD correctly predicts a gene that starts at position 148 and a frameshift between positions 911 and 912. The frameshift expectation and the ‘mean’ prediction make clear the uncertainties on the frameshift and START positions.
Similar articles
- EUGENE'HOM: A generic similarity-based gene finder using multiple homologous sequences.
Foissac S, Bardou P, Moisan A, Cros MJ, Schiex T. Foissac S, et al. Nucleic Acids Res. 2003 Jul 1;31(13):3742-5. doi: 10.1093/nar/gkg586. Nucleic Acids Res. 2003. PMID: 12824408 Free PMC article. - GeneTack database: genes with frameshifts in prokaryotic genomes and eukaryotic mRNA sequences.
Antonov I, Baranov P, Borodovsky M. Antonov I, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D152-6. doi: 10.1093/nar/gks1062. Epub 2012 Nov 17. Nucleic Acids Res. 2013. PMID: 23161689 Free PMC article. - OrfPredictor: predicting protein-coding regions in EST-derived sequences.
Min XJ, Butler G, Storms R, Tsang A. Min XJ, et al. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W677-80. doi: 10.1093/nar/gki394. Nucleic Acids Res. 2005. PMID: 15980561 Free PMC article. - Fast masking of repeated primer binding sites in eukaryotic genomes.
Andreson R, Kaplinski L, Remm M. Andreson R, et al. Methods Mol Biol. 2007;402:201-18. doi: 10.1007/978-1-59745-528-2_10. Methods Mol Biol. 2007. PMID: 17951797 Review. - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
Cited by
- An approach for searching insertions in bacterial genes leading to the phase shift of triplet periodicity.
Korotkova MA, Kudryashov NA, Korotkov EV. Korotkova MA, et al. Genomics Proteomics Bioinformatics. 2011 Oct;9(4-5):158-70. doi: 10.1016/S1672-0229(11)60019-3. Genomics Proteomics Bioinformatics. 2011. PMID: 22196359 Free PMC article. - Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads.
Darrasse A, Carrère S, Barbe V, Boureau T, Arrieta-Ortiz ML, Bonneau S, Briand M, Brin C, Cociancich S, Durand K, Fouteau S, Gagnevin L, Guérin F, Guy E, Indiana A, Koebnik R, Lauber E, Munoz A, Noël LD, Pieretti I, Poussier S, Pruvost O, Robène-Soustrade I, Rott P, Royer M, Serres-Giardi L, Szurek B, van Sluys MA, Verdier V, Vernière C, Arlat M, Manceau C, Jacques MA. Darrasse A, et al. BMC Genomics. 2013 Nov 6;14:761. doi: 10.1186/1471-2164-14-761. BMC Genomics. 2013. PMID: 24195767 Free PMC article. - Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes.
Rymen B, Fiorani F, Kartal F, Vandepoele K, Inzé D, Beemster GT. Rymen B, et al. Plant Physiol. 2007 Mar;143(3):1429-38. doi: 10.1104/pp.106.093948. Epub 2007 Jan 5. Plant Physiol. 2007. PMID: 17208957 Free PMC article. - The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533.
Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet AC, Zwahlen MC, Rouvet M, Altermann E, Barrangou R, Mollet B, Mercenier A, Klaenhammer T, Arigoni F, Schell MA. Pridmore RD, et al. Proc Natl Acad Sci U S A. 2004 Feb 24;101(8):2512-7. doi: 10.1073/pnas.0307327101. Proc Natl Acad Sci U S A. 2004. PMID: 14983040 Free PMC article. - Genome Sequences of Three Atypical Xanthomonas campestris pv. campestris Strains, CN14, CN15, and CN16.
Bolot S, Roux B, Carrere S, Jiang BL, Tang JL, Arlat M, Noël LD. Bolot S, et al. Genome Announc. 2013 Jul 11;1(4):e00465-13. doi: 10.1128/genomeA.00465-13. Genome Announc. 2013. PMID: 23846270 Free PMC article.
References
- Salanoubat M., Genin,S., Artiguenave,F., Gouzy,J., Mangenot,S., Arlat,M., Billault,A., Brottier,P., Camus,J., Cattolico,L. et al. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum. Nature, 415, 497–502. - PubMed
- Galibert F., Finan,T.M., Long,S.R., Puhler,A., Abola,P., Ampe,F., Barloy-Hubler,F., Barnett,M.J., Becker,A., Boistard,P. et al. (2001) The composite genome of the legume symbiont Sinorhizobium meliloti. Science, 293, 668–672. - PubMed
- Serra M., Turner,D. and Freier,S. (1995) Predicting thermodynamic properties of RNA. Methods Enzymol., 259, 243–261. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous