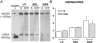Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles - PubMed (original) (raw)
Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles
A Garnier et al. J Physiol. 2003.
Abstract
Congestive heart failure (CHF) induces alterations in energy metabolism and mitochondrial function that span cardiac as well as skeletal muscles. Whether these defects originate from altered mitochondrial DNA copy number and/or mitochondrial gene transcription is not known at present, nor are the factors that control mitochondrial capacity in different muscle types completely understood. We used an experimental model of CHF induced by aortic banding in the rat and investigated mitochondrial respiration and enzyme activity of biochemical mitochondrial markers in cardiac, slow and fast skeletal muscles. We quantified mitochondrial DNA (mtDNA), expression of nuclear (COX IV) and mitochondrial (COX I) encoded cytochrome c oxidase subunits as well as nuclear factors involved in mitochondrial biogenesis and in the necessary coordinated interplay between nuclear and mitochondrial genomes in health and CHF. CHF induced a decrease in oxidative capacity and mitochondrial enzyme activities with a parallel decrease in the mRNA level of COX I and IV, but no change in mtDNA content. The expression of the peroxisome proliferator activated receptor gamma co-activator 1 alpha (PGC-1 alpha) gene was downregulated in CHF, as well as nuclear respiratory factor 2 and mitochondrial transcription factor A, which act downstream from PGC-1 alpha. Most interestingly, only the level of PGC-1 alpha expression was strongly correlated with muscle oxidative capacity in cardiac and skeletal muscles, both in healthy and CHF rats. Mitochondrial gene transcription is reduced in CHF, and PGC-1 alpha appears as a potential modulator of muscle oxidative capacity under these experimental conditions.
Figures
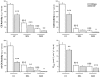
Figure 1. Enzymatic activities and mitochondrial function in left ventricle (LV), soleus (SOL) and gastrocnemius (GAS) of sham-operated and CHF rats
CS, citrate synthase; COX, cytochrome c oxidase; mi-CK, mitochondrial creatine kinase and  , maximal respiration rate. Values are means ±
, maximal respiration rate. Values are means ±
s.e.m
. (n = 11). *P < 0.05; **P < 0.01; ***,†††P < 0.001 versus respective Sham group (*) or versus Sham LV (†).
Figure 2. Southern blot analysis of total DNA from left ventricle (LV), soleus (SOL) and gastrocnemius (GAS) of sham-operated (S) and CHF rats
A, representative Southern blots for each muscle type with signals resulting from mitochondrial DNA (mtDNA) and nuclear DNA (nDNA). A positive control corresponding to a plasmid carrying sequence complementary to the mitochondrial probe was included and provided a signal at 4365 bp. B, mtDNA-to-nDNA ratio was determined after quantification of each signal. Values are means ±
s.e.m
. (n = 11).
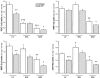
Figure 4. Real-time quantitative RT-PCR analysis of mRNA expression of peroxisome proliferator activated receptor gamma co-activator 1α (PGC-1α), nuclear respiratory factor 1 (NRF-1), nuclear respiratory factor 2 DNA-binding subunit α (NRF-2α) and mitochondrial transcription factor A (mtTFA) in left ventricle (LV), soleus (SOL) and gastrocnemius (GAS) of sham-operated and CHF rats
Results are given as means ±
s.e.m
. of values normalized to β-actin transcription and multiplied by total RNA (mg wet weight)−1. *,†P < 0.05; **,††P < 0.01; ***,†††P < 0.001 versus respective Sham group (*) or versus Sham LV (†).
Figure 3. Real-time quantitative RT-PCR analysis of mRNA expression of mitochondrial DNA (mtDNA)-encoded cytochrome c oxidase subunit I (COX I) and nuclear DNA (nDNA)-encoded cytochrome c oxidase subunit IV (COX IV) in left ventricle (LV), soleus (SOL) and gastrocnemius (GAS) of sham-operated and CHF rats
A, results are given as means ±
s.e.m
. of values normalized to β-actin transcription and multiplied by total RNA (mg wet weight)−1. *,†P < 0.05; **,††P < 0.01; ***,†††P < 0.001 versus respective Sham group (*) or versus Sham LV (†). B, correlations between the mRNA levels of COX I or COX IV and PGC-1α (peroxisome proliferator activated receptor gamma co-activator 1α). r, the correlation coefficient; P, the statistical significance.
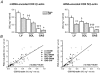
Figure 5. Correlations between the mRNA expression level of PGC-1α and enzymatic activities of citrate synthase (CS), cytochrome c oxidase (COX), mitochondrial creatine kinase (mi-CK) or maximal respiration rate () in left ventricle (LV), soleus (SOL) and gastrocnemius (GAS) of sham-operated and CHF rats
r, the correlation coefficient; P, the statistical significance.
Similar articles
- Methionine restriction effects on mitochondrial biogenesis and aerobic capacity in white adipose tissue, liver, and skeletal muscle of F344 rats.
Perrone CE, Mattocks DA, Jarvis-Morar M, Plummer JD, Orentreich N. Perrone CE, et al. Metabolism. 2010 Jul;59(7):1000-11. doi: 10.1016/j.metabol.2009.10.023. Epub 2010 Jan 4. Metabolism. 2010. PMID: 20045141 - The deacetylase enzyme SIRT1 is not associated with oxidative capacity in rat heart and skeletal muscle and its overexpression reduces mitochondrial biogenesis.
Gurd BJ, Yoshida Y, Lally J, Holloway GP, Bonen A. Gurd BJ, et al. J Physiol. 2009 Apr 15;587(Pt 8):1817-28. doi: 10.1113/jphysiol.2008.168096. Epub 2009 Feb 23. J Physiol. 2009. PMID: 19237425 Free PMC article. - Regional absence of mitochondria causing energy depletion in the myocardium of muscle LIM protein knockout mice.
van den Bosch BJ, van den Burg CM, Schoonderwoerd K, Lindsey PJ, Scholte HR, de Coo RF, van Rooij E, Rockman HA, Doevendans PA, Smeets HJ. van den Bosch BJ, et al. Cardiovasc Res. 2005 Feb 1;65(2):411-8. doi: 10.1016/j.cardiores.2004.10.025. Cardiovasc Res. 2005. PMID: 15639480 - Mitochondrial oxidative stress and dysfunction in myocardial remodelling.
Tsutsui H, Kinugawa S, Matsushima S. Tsutsui H, et al. Cardiovasc Res. 2009 Feb 15;81(3):449-56. doi: 10.1093/cvr/cvn280. Epub 2008 Oct 14. Cardiovasc Res. 2009. PMID: 18854381 Review. - Skeletal muscle disorders in heart failure.
Lunde PK, Sjaastad I, Schiøtz Thorud HM, Sejersted OM. Lunde PK, et al. Acta Physiol Scand. 2001 Mar;171(3):277-94. doi: 10.1046/j.1365-201x.2001.00830.x. Acta Physiol Scand. 2001. PMID: 11412140 Review.
Cited by
- Rescue of Heart Failure by Mitochondrial Recovery.
Marquez J, Lee SR, Kim N, Han J. Marquez J, et al. Int Neurourol J. 2016 Mar;20(1):5-12. doi: 10.5213/inj.1632570.285. Epub 2016 Mar 24. Int Neurourol J. 2016. PMID: 27032551 Free PMC article. Review. - The Complex Interplay between Mitochondria, ROS and Entire Cellular Metabolism.
Kuznetsov AV, Margreiter R, Ausserlechner MJ, Hagenbuchner J. Kuznetsov AV, et al. Antioxidants (Basel). 2022 Oct 8;11(10):1995. doi: 10.3390/antiox11101995. Antioxidants (Basel). 2022. PMID: 36290718 Free PMC article. Review. - Early combined treatment with sildenafil and adipose-derived mesenchymal stem cells preserves heart function in rat dilated cardiomyopathy.
Lin YC, Leu S, Sun CK, Yen CH, Kao YH, Chang LT, Tsai TH, Chua S, Fu M, Ko SF, Wu CJ, Lee FY, Yip HK. Lin YC, et al. J Transl Med. 2010 Sep 26;8:88. doi: 10.1186/1479-5876-8-88. J Transl Med. 2010. PMID: 20868517 Free PMC article. - Defective DNA replication impairs mitochondrial biogenesis in human failing hearts.
Karamanlidis G, Nascimben L, Couper GS, Shekar PS, del Monte F, Tian R. Karamanlidis G, et al. Circ Res. 2010 May 14;106(9):1541-8. doi: 10.1161/CIRCRESAHA.109.212753. Epub 2010 Mar 25. Circ Res. 2010. PMID: 20339121 Free PMC article. - The impact of thyroid hormone dysfunction on ischemic heart disease.
von Hafe M, Neves JS, Vale C, Borges-Canha M, Leite-Moreira A. von Hafe M, et al. Endocr Connect. 2019 May 1;8(5):R76-R90. doi: 10.1530/EC-19-0096. Endocr Connect. 2019. PMID: 30959486 Free PMC article. Review.
References
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. - PubMed
- Bergeron R, Ren JM, Cadman KS, Moore IK, Perret P, Pypaert M, Young LH, Semenkovich CF, Shulman GI. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Met. 2001;281:E1340–1346. - PubMed
- Cook SA, Matsui T, Li L, Rosenzweig A. Transcriptional effects of chronic Akt activation in the heart. J Biol Chem. 2002;277:22528–22533. - PubMed
- Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J Mol Biol. 1995;249:11–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical