Structural studies of the interaction of S-adenosylmethionine with the [4Fe-4S] clusters in biotin synthase and pyruvate formate-lyase activating enzyme - PubMed (original) (raw)
Structural studies of the interaction of S-adenosylmethionine with the [4Fe-4S] clusters in biotin synthase and pyruvate formate-lyase activating enzyme
Michele M Cosper et al. Protein Sci. 2003 Jul.
Abstract
The diverse reactions catalyzed by the radical-SAM superfamily of enzymes are thought to proceed via a set of common mechanistic steps, key among which is the reductive cleavage of S-adenosyl-L-methionine (SAM) by a reduced [4Fe-4S] cluster to generate an intermediate deoxyadenosyl radical. A number of spectroscopic studies have provided evidence that SAM interacts directly with the [4Fe-4S] clusters in several of the radical-SAM enzymes; however, the molecular mechanism for the reductive cleavage has yet to be elucidated. Selenium X-ray absorption spectroscopy (Se-XAS) was used previously to provide evidence for a close interaction between the Se atom of selenomethionine (a cleavage product of Se-SAM) and an Fe atom of the [4Fe-4S] cluster of lysine-2,3-aminomutase (KAM). Here, we utilize the same approach to investigate the possibility of a similar interaction in pyruvate formate-lyase activating enzyme (PFL-AE) and biotin synthase (BioB), two additional members of the radical-SAM superfamily. The results show that the latter two enzymes do not exhibit the same Fe-Se interaction as was observed in KAM, indicating that the methionine product of reductive cleavage of SAM does not occupy a well-defined site close to the cluster in PFL-AE and BioB. These results are interpreted in terms of the differences among these enzymes in their use of SAM as either a cofactor or a substrate.
Figures
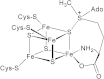
Figure 1.
Proposed mode of interaction of SAM with the [4Fe-4S] cluster of PFL-AE. The model is based on electron-nuclear double resonance and Mössbauer studies (Krebs et al. 2002; Walsby et al. 2002a,b).
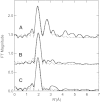
Figure 2.
Fourier transforms (over k = 2–12.5 Å−1) for (A) lysine 2,3-aminomutase incubated with SeMet, 5′deoxyadenosine, and didehydrolysine (solid line) and the calculated spectra for Se-C,Fe (Cosper et al. 2000); (B) PFL-AE [4Fe-4S]2+ incubated with SeMet and PFL (solid line) and the calculated spectra for Se-C2 (broken line; Fit 2, Table 1▶); and (C) BioB incubated with SeMet, d-biotin, and 5′deoxyadenosine (solid line) and the calculated spectra for Se-C2 (broken line; Fit 6, Table 1▶).
Similar articles
- Mechanism of Radical Initiation in the Radical S-Adenosyl-l-methionine Superfamily.
Broderick WE, Hoffman BM, Broderick JB. Broderick WE, et al. Acc Chem Res. 2018 Nov 20;51(11):2611-2619. doi: 10.1021/acs.accounts.8b00356. Epub 2018 Oct 15. Acc Chem Res. 2018. PMID: 30346729 Free PMC article. Review. - Coordination and mechanism of reversible cleavage of S-adenosylmethionine by the [4Fe-4S] center in lysine 2,3-aminomutase.
Chen D, Walsby C, Hoffman BM, Frey PA. Chen D, et al. J Am Chem Soc. 2003 Oct 1;125(39):11788-9. doi: 10.1021/ja036120z. J Am Chem Soc. 2003. PMID: 14505379 - The 4Fe-4S cluster in reconstituted biotin synthase binds S-adenosyl-L-methionine.
Cosper MM, Jameson GN, Davydov R, Eidsness MK, Hoffman BM, Huynh BH, Johnson MK. Cosper MM, et al. J Am Chem Soc. 2002 Nov 27;124(47):14006-7. doi: 10.1021/ja0283044. J Am Chem Soc. 2002. PMID: 12440894 - Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active 4Fe-4S cluster of pyruvate formate-lyase activating enzyme.
Walsby CJ, Hong W, Broderick WE, Cheek J, Ortillo D, Broderick JB, Hoffman BM. Walsby CJ, et al. J Am Chem Soc. 2002 Mar 27;124(12):3143-51. doi: 10.1021/ja012034s. J Am Chem Soc. 2002. PMID: 11902903 - Pyruvate formate-lyase activating enzyme: elucidation of a novel mechanism for glycyl radical formation.
Buis JM, Broderick JB. Buis JM, et al. Arch Biochem Biophys. 2005 Jan 1;433(1):288-96. doi: 10.1016/j.abb.2004.09.028. Arch Biochem Biophys. 2005. PMID: 15581584 Review.
Cited by
- Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins.
Nicolet Y, Amara P, Mouesca JM, Fontecilla-Camps JC. Nicolet Y, et al. Proc Natl Acad Sci U S A. 2009 Sep 1;106(35):14867-71. doi: 10.1073/pnas.0904385106. Epub 2009 Aug 17. Proc Natl Acad Sci U S A. 2009. PMID: 19706452 Free PMC article. - Structural insights into radical generation by the radical SAM superfamily.
Vey JL, Drennan CL. Vey JL, et al. Chem Rev. 2011 Apr 13;111(4):2487-506. doi: 10.1021/cr9002616. Epub 2011 Mar 3. Chem Rev. 2011. PMID: 21370834 Free PMC article. Review. No abstract available. - Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme.
Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Berkovitch F, et al. Science. 2004 Jan 2;303(5654):76-9. doi: 10.1126/science.1088493. Science. 2004. PMID: 14704425 Free PMC article. - Structural diversity in the AdoMet radical enzyme superfamily.
Dowling DP, Vey JL, Croft AK, Drennan CL. Dowling DP, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1178-95. doi: 10.1016/j.bbapap.2012.04.006. Epub 2012 Apr 28. Biochim Biophys Acta. 2012. PMID: 22579873 Free PMC article. Review. - Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes.
Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. Layer G, et al. EMBO J. 2003 Dec 1;22(23):6214-24. doi: 10.1093/emboj/cdg598. EMBO J. 2003. PMID: 14633981 Free PMC article.
References
- Baldet, P., Alban, C., and Douce, R. 1997. Biotin synthesis in higher plants: Purification and characterization of bioB gene product equivalent from Arabidopsis thaliana overexpressed in Escherichia coli and its subcellular localization in pea leaf cells. FEBS Lett. 419 206–210. - PubMed
- Beinert, H. 2000. Iron-sulfur proteins: Ancient structures, still full of surprises. J. Biol. Inorg. Chem. 5 2–15. - PubMed
- Beinert, H., Holm, R.H., and Münck, E. 1997. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 277 653–659. - PubMed
- Broderick, J.B., Duderstadt, R.E., Fernandez, D.C., Wojtuszewski, K., Henshaw, T.F., and Johnson, M.K. 1997. Pyruvate formate-lyase activating enzyme is an iron-sulfur protein. J. Am. Chem. Soc. 119 7396–7397.
- Broderick, J.B., Henshaw, T.F., Cheek, J., Wojtuszewski, K., Smith, S.R., Trojan, M.R., McGhan, R.M., Kopf, A., Kibbey, M., and Broderick, W.E. 2000. Pyruvate formate-lyase-activating enzyme: Strictly anaerobic isolation yields active enzyme containing a [3Fe-4S]+ cluster. Biochem. Biophys. Res. Commun. 269 451–456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK 59730/DK/NIDDK NIH HHS/United States
- R01 GM054608/GM/NIGMS NIH HHS/United States
- GM 62542/GM/NIGMS NIH HHS/United States
- GM 54608/GM/NIGMS NIH HHS/United States
- GM 42025/GM/NIGMS NIH HHS/United States
- R01 GM042025/GM/NIGMS NIH HHS/United States
- F32 DK059730/DK/NIDDK NIH HHS/United States
- R01 GM062542/GM/NIGMS NIH HHS/United States
- R29 GM054608/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases