Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes - PubMed (original) (raw)
Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes
Angélica Figueroa et al. Mol Cell Biol. 2003 Jul.
Abstract
In this report, we investigate the role of the RNA-binding protein HuR during skeletal myogenesis. At the onset of myogenesis in differentiating C2C12 myocytes and in vivo in regenerating mouse muscle, HuR cytoplasmic abundance increased dramatically, returning to a predominantly nuclear presence upon completion of myogenesis. mRNAs encoding key regulators of myogenesis-specific transcription (myogenin and MyoD) and cell cycle withdrawal (p21), bearing AU-rich regions, were found to be targets of HuR in a differentiation-dependent manner. Accordingly, mRNA half-lives were highest during differentiation, declining when differentiation was completed. Importantly, HuR-overexpressing C2C12 cells displayed increased target mRNA expression and half-life and underwent precocious differentiation. Our findings underscore a critical function for HuR during skeletal myogenesis linked to HuR's coordinate regulation of muscle differentiation genes.
Figures
FIG. 1.
Myogenin expression and mRNA stability are altered during differentiation of C2C12 muscle cells. (A) Asynchronously growing C2C12 cells (top) were subjected to differentiation using ITS. After 120 h (5 days) in continuous presence of ITS, cultures were fully differentiated and displayed abundant myotubes (bottom). (B) Top, Northern blot analysis of myogenin expression in C2C12 cells undergoing differentiation. 18S rRNA signals served to control for RNA loading. The graph depicts myogenin mRNA signal intensities (relative to 18S signals) shown as the means ± standard errors of the means from three independent Northern blot analyses. Bottom, Western blot analysis of myogenin expression in C2C12 cells undergoing differentiation for the times indicated. All lanes on the Western blot were loaded equally. (C) Nuclear run-on analysis to monitor transcription in C2C12 cells undergoing differentiation for the time periods shown. Nascent, radiolabeled RNA was hybridized to 1 μg of PCR-generated and slot blotted cDNAs; pUC19 plasmid was included as control for nonspecific hybridization. (D) Assessment of myogenin mRNA stability during C2C12 cell differentiation. mRNA half-lives were calculated after treatment of cells with 2.5 μg of ActD/ml and preparation of RNA at the times indicated; myogenin mRNA signals were measured, normalized to 18S rRNA signals, and plotted on a logarithmic scale. Horizontal dashed lines, 50% of untreated. Data represent the means ± standard deviations from three experiments. Bar graphs depict mRNA half-life values during C2C12 differentiation.
FIG. 2.
Half-lives of mRNAs encoding the MyoD and p21 muscle differentiation-associated genes change during C2C12 myocyte differentiation. (A) Northern blot analysis of MyoD, p21, and GAPDH expression in C2C12 cells undergoing differentiation for the times shown. The graph depicts the intensities of mRNAs encoding p21 and MyoD (relative to 18S), shown as the means ± standard errors of the means from three independent Northern blot analyses. (B) The stability of mRNAs encoding MyoD, p21, and GAPDH was calculated as explained in the legend for Fig. 1D.
FIG.3.
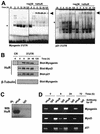
In vitro binding of HuR to the myogenin 3′ UTR. mRNAs encoding myogenin, MyoD, and p21 form complexes with C2C12 cytoplasmic proteins in a time-dependent fashion. (A) Schematic representation of the mRNAs encoding myogenin, MyoD, and p21, and various transcripts corresponding to the coding region and 3′ UTR used in this study. Boxes, RNA sequences of the 3′ UTR transcripts used; the p21 3′ UTR probe was previously reported (20). (B) REMSA detection of RNA-protein complexes. PCR-amplified cDNA fragments were used as templates to synthesize 32P-radiolabeled RNA probes. Cytoplasmic and nuclear fractions (10 μg each) prepared at the times indicated during C2C12 differentiation were assayed for binding to the indicated RNA probes. Complexes were resolved on 7% native polyacrylamide gels that were subsequently dried and visualized with a PhosphorImager. Brackets indicate inducible RNA-protein complexes. f (free probe), radiolabeled RNA incubated only with RNase T1. (C) Cytoplasmic fractions prepared from differentiating C2C12 cells were incubated with myogenin, MyoD, and p21 3′ UTR transcripts, and the resulting radiolabeled complexes were digested with RNase T1 and subjected to UV cross-linking and electrophoresis through sodium dodecyl sulfate-containing 15% polyacrylamide gels. Upon drying of gels, signals were visualized using a PhosphorImager. Numbers denote sizes (in kDa) of molecular weight markers (MWM). Open arrowheads, radiolabeled complexes; black arrowheads, predicted position of HuR. (D) An RNase T1 selection assay was carried out using myogenin transcripts and 10 nM either GST or GST-HuR. T1, digestions with RNase T1 alone. Arrowhead, protected RNA fragments. (E) Agarose gel retardation assays using myogenin 3′ UTR and the indicated concentrations of either GST or GST-HuR. Arrowhead, protected RNA fragments.
FIG. 4.
Endogenous HuR forms differentiation-dependent complexes with recombinant and endogenous transcripts encoding myogenin, MyoD, and p21. (A) Complexes forming with cytoplasmic lysates from C2C12 cells undergoing differentiation for the times indicated and the radiolabeled transcripts shown were assayed for their ability to be supershifted by 0.5 μg of either anti-HuR antibody (+αHuR) or a control antibody recognizing the mitogen-activated protein kinase p38 (+αp38). Arrowheads, HuR-containing supershifted complexes. (B) Biotinylated transcripts corresponding to the 3′ UTRs of each mRNA, as well as the myogenin coding region (CR) (negative control), were incubated with cytoplasmic C2C12 lysates prepared at the times shown after induction of differentiation. Complexes were pulled down using streptavidin-conjugated magnetic beads, and HuR abundance was analyzed by Western blotting. (C) HuR in C2C12 lysates after IP using either anti-HuR antibodies (3A2) or isotype-matched IgG1 was assessed by Western blot analysis (WB) of the IP material. (D) The presence of HuR target mRNAs encoding myogenin, MyoD, and p21 was assessed by RT-PCR analysis of IP material obtained using either anti-HuR or control IgG1 antibodies.
FIG. 5.
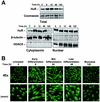
Effect of C2C12 cell differentiation on the subcellular localization of HuR. (A) Western blot analysis of HuR levels in cytoplasmic (10 μg), nuclear (10 μg), and whole-cell (20 μg) lysates prepared from C2C12 cells that were undergoing differentiation for the times indicated. (B) Immunofluorescence detection of HuR in C2C12 cells throughout the differentiation process. Magnification, ×40; zoom, close-up images.
FIG. 6.
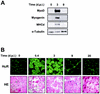
Cytoplasmic localization of HuR after induced injury in mouse muscle. (A) Muscle lysates from either intact (0 days postinjury [d.p.i.]) or cardiotoxin-injured muscles (3 and 9 d.p.i.) were prepared for Western blot analysis to assess the expression of regeneration markers MyoD, myogenin, and MHCd, as well as loading control α-tubulin. (B) Immunofluorescent detection of HuR in frozen muscle sections prepared at the indicated d.p.i. Sections were stained with HE to monitor adequate progression of the degeneration-regeneration process. Magnification, ×60.
FIG. 7.
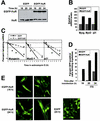
Overexpression of HuR increases the expression of differentiation-associated genes and accelerates myotube formation. C2C12 cells were transiently transfected with plasmids expressing either EGFP or EGFP-HuR. (A) Western blot analysis of HuR expression using whole-cell lysates. (B) Quantitation of Northern blot signals to measure steady-state mRNA levels 16 h after transfection of either pEGFP or pEGFP-HuR (10 h in the presence of ITS). (C) mRNA half-lives assessed 16 h after transfection (10 h in the presence of ITS), as explained in the legend for Fig. 1D; data are the means of two similar experiments. (D) Following C2C12 transfection, the percentage of EGFP-positive cells forming syncytia (two or more fused myocytes) was assessed in each transfection group. (E) Representative fields depicting early myotube formation in EGFP-HuR-overexpressing C2C12 cells that were transiently transfected as described above. Cultures were examined by phase contrast (data not shown) and fluorescence microscopy; syncytia are indicated with a white discontinuous line. Magnification, ×40.
Similar articles
- RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation.
van der Giessen K, Di-Marco S, Clair E, Gallouzi IE. van der Giessen K, et al. J Biol Chem. 2003 Nov 21;278(47):47119-28. doi: 10.1074/jbc.M308889200. Epub 2003 Aug 27. J Biol Chem. 2003. PMID: 12944397 - Involvement of transportin 2-mediated HuR import in muscle cell differentiation.
van der Giessen K, Gallouzi IE. van der Giessen K, et al. Mol Biol Cell. 2007 Jul;18(7):2619-29. doi: 10.1091/mbc.e07-02-0167. Epub 2007 May 2. Mol Biol Cell. 2007. PMID: 17475777 Free PMC article. - Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation.
Cammas A, Sanchez BJ, Lian XJ, Dormoy-Raclet V, van der Giessen K, López de Silanes I, Ma J, Wilusz C, Richardson J, Gorospe M, Millevoi S, Giovarelli M, Gherzi R, Di Marco S, Gallouzi IE. Cammas A, et al. Nat Commun. 2014 Jun 27;5:4190. doi: 10.1038/ncomms5190. Nat Commun. 2014. PMID: 24969639 Free PMC article. - HuR and myogenesis: being in the right place at the right time.
von Roretz C, Beauchamp P, Di Marco S, Gallouzi IE. von Roretz C, et al. Biochim Biophys Acta. 2011 Sep;1813(9):1663-7. doi: 10.1016/j.bbamcr.2011.01.036. Epub 2011 Feb 20. Biochim Biophys Acta. 2011. PMID: 21315776 Review. - Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis.
Zammit PS. Zammit PS. Semin Cell Dev Biol. 2017 Dec;72:19-32. doi: 10.1016/j.semcdb.2017.11.011. Epub 2017 Nov 15. Semin Cell Dev Biol. 2017. PMID: 29127046 Review.
Cited by
- Posttranscriptional regulation by RNA-binding proteins during epithelial-to-mesenchymal transition.
Aparicio LA, Abella V, Valladares M, Figueroa A. Aparicio LA, et al. Cell Mol Life Sci. 2013 Dec;70(23):4463-77. doi: 10.1007/s00018-013-1379-0. Epub 2013 May 29. Cell Mol Life Sci. 2013. PMID: 23715860 Free PMC article. Review. - Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay.
Hausburg MA, Doles JD, Clement SL, Cadwallader AB, Hall MN, Blackshear PJ, Lykke-Andersen J, Olwin BB. Hausburg MA, et al. Elife. 2015 Mar 27;4:e03390. doi: 10.7554/eLife.03390. Elife. 2015. PMID: 25815583 Free PMC article. - Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells.
Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Lee JE, et al. PLoS One. 2010 Jun 21;5(6):e11201. doi: 10.1371/journal.pone.0011201. PLoS One. 2010. PMID: 20574513 Free PMC article. - Overexpression of the RNA binding protein HuR impairs tumor growth in triple negative breast cancer associated with deficient angiogenesis.
Gubin MM, Calaluce R, Davis JW, Magee JD, Strouse CS, Shaw DP, Ma L, Brown A, Hoffman T, Rold TL, Atasoy U. Gubin MM, et al. Cell Cycle. 2010 Aug 15;9(16):3337-46. doi: 10.4161/cc.9.16.12711. Epub 2010 Aug 17. Cell Cycle. 2010. PMID: 20724828 Free PMC article. - Staufen1 inhibits MyoD translation to actively maintain muscle stem cell quiescence.
de Morrée A, van Velthoven CTJ, Gan Q, Salvi JS, Klein JDD, Akimenko I, Quarta M, Biressi S, Rando TA. de Morrée A, et al. Proc Natl Acad Sci U S A. 2017 Oct 24;114(43):E8996-E9005. doi: 10.1073/pnas.1708725114. Epub 2017 Oct 9. Proc Natl Acad Sci U S A. 2017. PMID: 29073096 Free PMC article.
References
- Atasoy, U., J. Watson, D. Patel, and J. D. Keene. 1998. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 111:3145-3156. - PubMed
- Beckel-Mitchener, A. C., A. Miera, R. Keller, and N. I. Perrone-Bizzozero. 2002. Poly(A) tail length-dependent stabilization of GAP-43 mRNA by the RNA-binding protein HuD. J. Biol. Chem. 277:27996-28002. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous