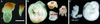Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability - PubMed (original) (raw)
Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability
Ellen E McCarthy et al. Mol Cell Biol. 2003 Jul.
Abstract
The BRCA1 tumor suppressor has been implicated in many cellular pathways, but the mechanisms by which it suppresses tumor formation are not fully understood. In vivo BRCA1 forms a heterodimeric complex with the related BARD1 protein, and its enzymatic activity as a ubiquitin ligase is largely dependent upon its interaction with BARD1. To explore the genetic relationship between BRCA1 and BARD1, we have examined the phenotype of Bard1-null mice. These mice become developmentally retarded and die between embryonic day 7.5 (E7.5) and E8.5. Embryonic lethality results from a severe impairment of cell proliferation that is not accompanied by increased apoptosis. In the absence of p53, the developmental defects associated with Bard1 deficiency are partly ameliorated, and the lethality of Bard1; p53-nullizygous mice is delayed until E9.5. This result, together with the increased chromosomal aneuploidy of Bard1 mutant cells, indicates a role for Bard1 in maintaining genomic stability. The striking similarities between the phenotypes of Bard1-null, Brca1-null, and double Bard1; Brca1-null mice provide strong genetic evidence that the developmental functions of Brca1 and Bard1 are mediated by the Brca1/Bard1 heterodimer.
Figures
FIG. 1.
Targeted disruption of the mouse Bard1 gene. (A) A partial restriction map of the genomic region encompassing Bard1 exon 1 (Ex1; black box) is shown on top, followed by a diagram of the targeting vector used for insertion of the hygromycin resistance-EGFP gene cassette (HygEGFP) into exon 1 of the mouse Bard1 gene (bottom). A diphtheria toxin A gene (DT) was also included in the targeting construct for negative selection. The wavy lines represent plasmid sequences. Relevant restriction sites are _Eco_RI (E), _Hin_dIII (H), _Pst_I (P), and _Sal_I (S). The 5′ flanking probe used for Southern analyses, the sizes of the endogenous and expected DNA fragments, and the locations of the PCR primers a, b, and c are also shown. hyg, hygromycin resistance gene. (B) Schematics of the neomycin resistance gene (neo) targeting vector and the recombined Bard1 locus are shown. (C) Southern analysis of ES cell DNA digested with _Pst_I to confirm gene targeting. Due to an additional _Pst_I site in the hygromycin selection marker, the 7.2-kb _Pst_I fragment detected in wild-type ES cells is reduced to 5.5 kb in properly targeted ES cells. (D) Southern analysis to identify the second targeting event in heterozygous ES cells. wt, wild type. (E) Representative PCR genotyping of E6.5 embryos with primers a, b, and c. PCR amplification was conducted on embryonic DNA from wild-type (lanes 1 and 2), Bard1+/− (lane 3), and _Bard1_−/− (lane 4) embryos. The PCR products derived from wild-type and mutant alleles are 347 and 422 bp, respectively.
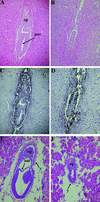
FIG. 2.
Development of _Bard1_−/− embryos. Hematoxylin- and eosin-stained sagittal sections of wild-type (A and E) and _Bard1_−/− (B and F) embryos at E6.5 (A and B) and E7.5 (E and F) are shown. (A) Normal (Bard1+/−) egg cylinder with proamniotic cavity (pac) and ectoplacental cone (ep). (B) Developmentally retarded Bard1 mutant embryo. (C and D) The proliferating cells of wild-type (C) and Bard1 mutant (D) E6.5 embryos are highlighted by BrdU labeling. Strong BrdU staining can be detected throughout the wild-type embryo (C), whereas the Bard1 mutant embryo has fewer BrdU-labeled nuclei (D). (E) E7.5 wild-type embryo postgastrulation with three distinct germ layers. am, amnion; y, yolk sac. (F) Bard1 homozygous mutant at the egg cylinder stage with prominent proamniotic cavity.
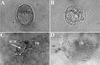
FIG. 3.
Impaired proliferation of _Bard1_−/− mutants in vitro. (A) Wild-type E3.5 blastocyst. (B) _Bard1_−/− E3.5 blastocysts. (C and D) Wild-type (C) and _Bard1_−/− (D) blastocyst outgrowths after 6 days in culture. Proliferation of both the ICM and the trophoblastic giant cells (TR) is evident in the wild-type embryo. In contrast, in the _Bard1_−/− mutant embryo, the ICM is completely missing and only the trophoblastic giant cells remain.
FIG. 4.
Gross morphologies of wild-type, single homozygous mutant, and double homozygous mutant embryos. (A) Normal (Bard1+/−) and mutant (_Bard1_−/−) embryos at E7.5. (B) A normal (Bard1+/−; Brca1+/−) E9.5 embryo is shown on the left. A Bard1; Brca1 (bd1/br1) double homozygous mutant embryo is shown in comparison to a single Bard1 (bd1) mutant (heterozygous for Brca1) and a single Brca1 (br1) nullizygote (wild type for Bard1) that exhibit similar extents of development. (C) A normal (Bard1+/+; _p53_−/−) E9.5 embryo on the right compared to a Bard1; p53 double homozygous mutant (_Bard1_−/−; _p53_−/−) E9.5 embryo on the left. Note that the Bard1; p53 double homozygous mutant E9.5 embryo has developed much further than any single _Bard1_−/− embryo. Scale bar, 300 μm (A) and 800 μm (B and C).
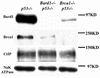
FIG. 5.
Reciprocal stability control of Bard1 and Brca1 proteins in vivo. Protein lysates of _Bard1+/+; p53_−/−, _Bard1_−/−; _p53_−/−, and _Brca1_−/−; _p53_−/− embryos were analyzed by Western blotting. Anti-NaK-ATPase was used as a loading control.
Similar articles
- The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression.
Shakya R, Szabolcs M, McCarthy E, Ospina E, Basso K, Nandula S, Murty V, Baer R, Ludwig T. Shakya R, et al. Proc Natl Acad Sci U S A. 2008 May 13;105(19):7040-5. doi: 10.1073/pnas.0711032105. Epub 2008 Apr 28. Proc Natl Acad Sci U S A. 2008. PMID: 18443292 Free PMC article. - The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation.
Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. Hashizume R, et al. J Biol Chem. 2001 May 4;276(18):14537-40. doi: 10.1074/jbc.C000881200. Epub 2001 Mar 6. J Biol Chem. 2001. PMID: 11278247 - The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity.
Baer R, Ludwig T. Baer R, et al. Curr Opin Genet Dev. 2002 Feb;12(1):86-91. doi: 10.1016/s0959-437x(01)00269-6. Curr Opin Genet Dev. 2002. PMID: 11790560 Review. - Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis.
Irminger-Finger I, Leung WC, Li J, Dubois-Dauphin M, Harb J, Feki A, Jefford CE, Soriano JV, Jaconi M, Montesano R, Krause KH. Irminger-Finger I, et al. Mol Cell. 2001 Dec;8(6):1255-66. doi: 10.1016/s1097-2765(01)00406-3. Mol Cell. 2001. PMID: 11779501 - The functions of breast cancer susceptibility gene 1 (BRCA1) product and its associated proteins.
Irminger-Finger I, Siegel BD, Leung WC. Irminger-Finger I, et al. Biol Chem. 1999 Feb;380(2):117-28. doi: 10.1515/BC.1999.019. Biol Chem. 1999. PMID: 10195418 Review.
Cited by
- BRCA1 Mutations in Cancer: Coordinating Deficiencies in Homologous Recombination with Tumorigenesis.
Krais JJ, Johnson N. Krais JJ, et al. Cancer Res. 2020 Nov 1;80(21):4601-4609. doi: 10.1158/0008-5472.CAN-20-1830. Epub 2020 Aug 3. Cancer Res. 2020. PMID: 32747362 Free PMC article. Review. - The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression.
Shakya R, Szabolcs M, McCarthy E, Ospina E, Basso K, Nandula S, Murty V, Baer R, Ludwig T. Shakya R, et al. Proc Natl Acad Sci U S A. 2008 May 13;105(19):7040-5. doi: 10.1073/pnas.0711032105. Epub 2008 Apr 28. Proc Natl Acad Sci U S A. 2008. PMID: 18443292 Free PMC article. - Functional characterization of full-length BARD1 strengthens its role as a tumor suppressor in neuroblastoma.
Cimmino F, Avitabile M, Lasorsa VA, Pezone L, Cardinale A, Montella A, Cantalupo S, Iolascon A, Capasso M. Cimmino F, et al. J Cancer. 2020 Jan 14;11(6):1495-1504. doi: 10.7150/jca.36164. eCollection 2020. J Cancer. 2020. PMID: 32047556 Free PMC article. - The BRCA1/BARD1 ubiquitin ligase and its substrates.
Witus SR, Stewart MD, Klevit RE. Witus SR, et al. Biochem J. 2021 Sep 30;478(18):3467-3483. doi: 10.1042/BCJ20200864. Biochem J. 2021. PMID: 34591954 Free PMC article. Review. - BARD1 Gene Polymorphisms Confer Nephroblastoma Susceptibility.
Fu W, Zhu J, Xiong SW, Jia W, Zhao Z, Zhu SB, Hu JH, Wang FH, Xia H, He J, Liu GC. Fu W, et al. EBioMedicine. 2017 Feb;16:101-105. doi: 10.1016/j.ebiom.2017.01.038. Epub 2017 Jan 31. EBioMedicine. 2017. PMID: 28161399 Free PMC article.
References
- Abbott, D. W., M. E. Thompson, C. Robinson-Benion, G. Tomlinson, R. A. Jensen, and J. T. Holt. 1999. BRCA1 expression restores radiation resistance in BRCA1-defective cancer cells through enhancement of transcription-coupled DNA repair. J. Biol. Chem. 274:18808-18812. - PubMed
- Baer, R., and T. Ludwig. 2002. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 12:86-91. - PubMed
- Bhattacharyya, A., U. S. Ear, B. H. Koller, R. R. Weichselbaum, and D. K. Bishop. 2000. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275:23899-23903. - PubMed
- Brodie, S. G., and C. X. Deng. 2001. BRCA1-associated tumorigenesis: what have we learned from knockout mice? Trends Genet. 17:S18-S22. - PubMed
- Brugarolas, J., and T. Jacks. 1997. Double indemnity: p53, BRCA and cancer. p53 mutation partially rescues developmental arrest in Brca1 and Brca2 null mice, suggesting a role for familial breast cancer genes in DNA damage repair. Nat. Med. 3:721-722. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous