PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis - PubMed (original) (raw)
PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis
Yoo-Sun Noh et al. Plant Cell. 2003 Jul.
Abstract
Proper control of the floral transition is critical for reproductive success in flowering plants. In Arabidopsis, FLOWERING LOCUS C (FLC) is a floral repressor upon which multiple floral regulatory pathways converge. Mutations in PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1) suppress the FLC-mediated delay of flowering as a result of the presence of FRIGIDA or of mutations in autonomous pathway genes. PIE1 is required for high levels of FLC expression in the shoot apex, but it is not required for FLC expression in roots. PIE1 is similar to ATP-dependent, chromatin-remodeling proteins of the ISWI and SWI2/SNF2 family. The role of PIE1 as an activator of FLC is consistent with the general role of ISWI and SWI2/SNF2 family genes as activators of gene expression. The pie1 mutation also causes early flowering in noninductive photoperiods independently of FLC; thus, PIE1 appears to be involved in multiple flowering pathways. PIE1 also plays a role in petal development, as revealed by the suppression of petal defects of the curly leaf mutant by the pie1 mutation.
Figures
Figure 1.
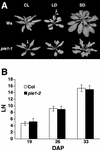
Flowering and Leaf Initiation Rate of pie1-1 Compared with the Wild Type. (A) Wild-type (Ws) and pie1-1 plants were grown under continuous light (CL), long days (LD), or short days (SD). Photographs were taken when flowering initiated and the inflorescence stem began to elongate. (B) The number of visible leaves (LN) in wild-type (Col) or pie1-2 plants grown in short days was scored at 19, 26, and 33 days after planting (DAP). Data shown are means ±
sd
of 12 plants per genotype.
Figure 2.
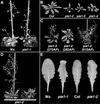
pie1 Mutant Phenotype in Different Genetic Backgrounds. (A) pie1-1 in the Ws background and wild-type Ws grown for 25 days in long days. (B) Wild-type Col and pie1-2, pie1-3, and pie1-4 in the Col background grown in long days until flowering had initiated. (C) The pleiotropic phenotype of Col pie1 alleles after the transition to flowering. A small fraction of pie1 mutant plants in Col showed some degree of primary inflorescence elongation (left); however, the majority of pie1 mutant plants displayed a more severe inhibition of primary inflorescence elongation and the development of numerous secondary inflorescences (middle), which led to a bushy phenotype in older plants (right). DAP, days after planting. (D) The genetic background influences the pie1 phenotype. Shown are representative pie1-2 plants from the F2 population obtained from a cross between pie1-2 in Col and Ws. Approximately three-quarters of the pie1-2 mutant plants displayed the more normal phenotype (left), whereas approximately one-quarter of the pie1-2 mutant plants displayed the more pleiotropic phenotype (right) characteristic of Col pie1 alleles. (E) Leaf phenotypes of pie1 mutants. Shown are the fifth rosette leaves from wild-type plants (Ws and Col) and pie1 mutants in the corresponding genetic backgrounds (pie1-1 and pie1-2, respectively) grown in short days.
Figure 3.
PIE1 Gene and Protein Structure. (A) Genomic arrangement of PIE1. The translation start and stop sites and the T-DNA insertion sites in pie1-1, pie1-2, pie1-3, and pie1-4 are indicated. Gray boxes indicate translated exons, and lines indicate introns or intergenic sequences. (B) Domains of PIE1. Domains predicted by the SMART (
http://smart.embl-heidelberg.de/
) program and the amino acid numbers of these domains are indicated. The two putative bipartite nuclear localization signals (NLS) at the N-terminal and C-terminal regions of PIE1 are KRQKTLEAPKEPRRPKT and KKRDLIVDTDEEKTSKK, respectively. (C) Sequence alignment of the SNF_N domain of PIE1 with Drosophila DOMINO A, human SRCAP, and yeast SWR1. Numerals indicate amino acid positions. (D) Sequence alignment of the HELICc domain. (E) Sequence alignment of the SANT domain. The PIE1 SANT domain was compared with the SANT domains from human ADA2, yeast ADA2, mouse N-CoR, yeast BAS1, human MYBA, yeast TFIIIB, yeast SWI3, and Drosophila ISWI. The two residues marked with stars have been shown to be important for the function of yeast ADA2 (Sterner et al., 2002).
Figure 4.
Suppression of _FLC_-Dependent Late Flowering by pie1. (A) Suppression of _FRI_-mediated late flowering by pie1. Representative plants of wild-type Ws and pie1-1 mutants homozygous for FRI grown in long days (LD) and short days (SD) are shown. Photographs were taken at the initiation of flowering. (B) Suppression of _ld_-mediated late flowering by pie1. Representative plants of wild-type Ws, the ld-2 mutant in the Ws background, and the ld-2 pie1-1 double mutant grown in long days are shown. Photographs were taken at the initiation of flowering. (C) Repression of _FRI_- or _ld_-mediated FLC activation by pie1. RNA was isolated from 10-day-old seedlings grown under continuous light. The blots were probed first with FLC and then reprobed with 18S ribosomal DNA (18S) as a loading control.
Figure 5.
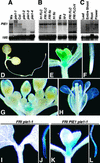
PIE1 Expression Pattern and the Effect of the pie1 Mutation on the FLC Expression Pattern. (A) PIE1 mRNA expression in wild-type and pie1 mutant alleles. RNA was isolated from 10-day-old seedlings of each genotype grown under continuous light (see [A] and [B]). The blots were probed first with PIE1 and then reprobed with 18S ribosomal DNA (18S) as a loading control (see [A], [B], and [C]). (B) The steady state PIE1 mRNA level is not regulated by FRI, FLC, LD, or vernalization. Genotype designations for FRI and FLC lines have been described previously (Michaels and Amasino, 2001). FRI FLC+V RNA was isolated from FRI FLC seedlings that had been vernalized for 40 days and then grown under continuous light for 7 days. (C) PIE1 mRNA expression in different tissues. Tissues were collected from adult plants grown in long days. (D) to (H) Histochemical GUS staining of transgenic Arabidopsis containing a PIE1 promoter:GUS fusion. (D) Seven-day-old whole seedling grown under continuous light. (E) and (F) Magnification of the shoot apical meristem region (E) and the root tip (F) of the seedling. (G) and (H) Inflorescence meristem region (G) and flower (H) from an adult plant grown in long days. (I) to (L) Histochemical GUS staining of transgenic Arabidopsis containing an FLC:GUS fusion. Shown are the shoot apical meristem region ([I] and [K]) and the root tip ([J] and [L]) of representative FRI pie1-1 homozygous and FRI PIE1 pie1-1 heterozygous seedlings grown for 7 days under continuous light.
Figure 6.
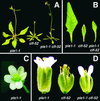
Effect of pie1 on the clf Phenotype. (A) pie1-1, clf-52, and pie1-1 clf-52 plants grown in long days to the flowering stage. (B) Fourth rosette leaves of pie1-1, clf-52, and pie1-1 clf-52 plants grown in long days. (C) pie1-1 flower with six petals. (D) Flowers of pie1-1, clf-52, and pie1-1 clf-52 plants grown in long days.
Similar articles
- Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications.
Richter R, Kinoshita A, Vincent C, Martinez-Gallegos R, Gao H, van Driel AD, Hyun Y, Mateos JL, Coupland G. Richter R, et al. PLoS Genet. 2019 Apr 4;15(4):e1008065. doi: 10.1371/journal.pgen.1008065. eCollection 2019 Apr. PLoS Genet. 2019. PMID: 30946745 Free PMC article. - Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z.
Deal RB, Topp CN, McKinney EC, Meagher RB. Deal RB, et al. Plant Cell. 2007 Jan;19(1):74-83. doi: 10.1105/tpc.106.048447. Epub 2007 Jan 12. Plant Cell. 2007. PMID: 17220196 Free PMC article. - Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development.
Choi K, Park C, Lee J, Oh M, Noh B, Lee I. Choi K, et al. Development. 2007 May;134(10):1931-41. doi: 10.1242/dev.001891. Development. 2007. PMID: 17470967 - The quest for florigen: a review of recent progress.
Corbesier L, Coupland G. Corbesier L, et al. J Exp Bot. 2006;57(13):3395-403. doi: 10.1093/jxb/erl095. Epub 2006 Oct 9. J Exp Bot. 2006. PMID: 17030536 Review. - Make hay when the sun shines: the role of MADS-box genes in temperature-dependant seasonal flowering responses.
Hemming MN, Trevaskis B. Hemming MN, et al. Plant Sci. 2011 Mar;180(3):447-53. doi: 10.1016/j.plantsci.2010.12.001. Epub 2010 Dec 14. Plant Sci. 2011. PMID: 21421391 Review.
Cited by
- Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Osakabe A, Takizawa Y, Horikoshi N, Hatazawa S, Negishi L, Sato S, Berger F, Kakutani T, Kurumizaka H. Osakabe A, et al. Nat Commun. 2024 Jul 11;15(1):5187. doi: 10.1038/s41467-024-49465-w. Nat Commun. 2024. PMID: 38992002 Free PMC article. - De novo assembly, gene annotation and marker development using Illumina paired-end transcriptome sequences in celery (Apium graveolens L.).
Fu N, Wang Q, Shen HL. Fu N, et al. PLoS One. 2013;8(2):e57686. doi: 10.1371/journal.pone.0057686. Epub 2013 Feb 28. PLoS One. 2013. PMID: 23469050 Free PMC article. - Histone Variants in the Specialization of Plant Chromatin.
Foroozani M, Holder DH, Deal RB. Foroozani M, et al. Annu Rev Plant Biol. 2022 May 20;73:149-172. doi: 10.1146/annurev-arplant-070221-050044. Epub 2022 Feb 15. Annu Rev Plant Biol. 2022. PMID: 35167758 Free PMC article. Review. - SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6.
March-Díaz R, García-Domínguez M, Florencio FJ, Reyes JC. March-Díaz R, et al. Plant Physiol. 2007 Feb;143(2):893-901. doi: 10.1104/pp.106.092270. Epub 2006 Dec 1. Plant Physiol. 2007. PMID: 17142478 Free PMC article. - CHR11, a chromatin-remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana.
Huanca-Mamani W, Garcia-Aguilar M, León-Martínez G, Grossniklaus U, Vielle-Calzada JP. Huanca-Mamani W, et al. Proc Natl Acad Sci U S A. 2005 Nov 22;102(47):17231-6. doi: 10.1073/pnas.0508186102. Epub 2005 Nov 14. Proc Natl Acad Sci U S A. 2005. PMID: 16286646 Free PMC article.
References
- Aasland, R., Stewart, A.F., and Gibson, T. (1996). The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21, 87–88. - PubMed
- Amedeo, P., Habu, Y., Afsar, K., Scheid, O.M., and Paszkowski, J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405, 203–206. - PubMed
- Aukerman, M.J., and Amasino, R.M. (1996). Molecular genetic analysis of flowering time in Arabidopsis. Semin. Cell Dev. Biol. 7, 427–433.
- Aukerman, M.J., Lee, I., Weigel, D., and Amasino, R.M. (1999). The Arabidopsis flowering-time gene LUMINIDEPENDENS is expressed primarily in regions of cell proliferation and encodes a nuclear protein that regulates LEAFY expression. Plant J. 18, 195–203. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous