The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis - PubMed (original) (raw)
The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis
Anup Dey et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Previous in vitro studies showed that the bromodomain binds to acetyllysines on histone tails, leading to the proposal that the domain is involved in deciphering the histone code. However, there is little in vivo evidence supporting the binding of bromodomains to acetylated chromatin in the native environment. Brd4 is a member of the BET family that carries two bromodomains. It associates with mitotic chromosomes, a feature characteristic of the family. Here, we studied the interaction of Brd4 with chromatin in living cells by photobleaching. Brd4 was mobile and interacted with chromatin with a rapid "on and off" mode of binding. This interaction required both bromodomains. Indicating a preferential interaction with acetylated chromatin, Brd4 became less mobile upon increased chromatin acetylation caused by a histone deacetylase inhibitor. Providing biochemical support, salt solubility of Brd4 was markedly reduced upon increased histone acetylation. This change also required both bromodomains. In peptide binding assays, Brd4 avidly bound to di- and tetraacetylated histone H4 and diacetylated H3, but weakly or not at all to mono- and unacetylated H3 and H4. By contrast, it did not bind to unacetylated H4 or H3. Further, Brd4 colocalized with acetylated H4 and H3 in noncentromeric regions of mitotic chromosomes. This colocalization also required both bromodomains. These observations indicate that Brd4 specifically recognizes acetylated histone codes, and this recognition is passed onto the chromatin of newly divided cells.
Figures
Fig. 2.
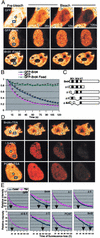
FLIP analysis of bromodomain point mutants. (A) Bromodomain sequence comparison. (B) Bromodomain point mutants tested in this study. (C) FLIP analysis of mutants tested without (control,Upper) or with (Lower) TSA treatment._T_1/2 of M5 and M6 was ≈120 s, whereas that of wild-type Brd4 and other mutants was ≈120 s. None of single point mutants showed a discernible difference in the FLIP pattern (data not shown).
Fig. 3.
Solubility of Brd4 in differential salt extraction. (A) Endogenous Brd4 was extracted with indicated concentrations of NaCl and detected by immunoblot. (B) Transfected GFP-Brd4 or GFP-Brd4 deletions were extracted and tested as in A. (C) Histone tail peptides tested for Brd4 binding (Top). Bound Brd4 was detected by immunoblot (Middle). TFIIB in unbound materials was detected to verify equal loading (Bottom).
Fig. 1.
FLIP analysis of GFP-Brd4. Murine P19 embryonal carcinoma cells expressing GFP-Brd4 or free GFP were photobleached in the circled area, and loss of fluorescence was monitored in the rectangle. (A) Distribution of GFP proteins before and after photobleaching. Bleaching of free GFP (Top) and GFP-Brd4 was performed with live (Middle) or fixed (Bottom) cells. (Bar indicates 3.6 μm.) (B) Quantification of fluorescence loss in A. Values represent the average of eight independent measurements ± SD. Arrows indicate the time (s) required for 50% fluorescence loss (_T_1/2). (C) Diagram of Brd4 deletions. Solid blocks (BD1 and BD2) mark bromodomains; shaded blocks mark the ET domain. (D) Distribution of indicated GFP proteins in cells pretreated with 50 ng/ml TSA for 4 h. (E) FLIP patterns for indicated GFP proteins without (control) or with TSA treatment.
Fig. 4.
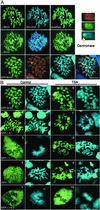
Chromosomal localization of Brd4 and Brd2. (A) Endogenous Brd4, Brd2, or acetylated H4 was visualized on mitotic chromosomes of P19 cells by indirect immunostaining. DNA was counterstained by Hoechst 33342. Arrows indicate centromeres where staining of Brd4, Brd2, or acetyl H4 was absent. (Bar indicates 8 μm.) (Upper Right) Enlargement of chromosome stained for acetyl H4 (red), Brd4 (green), and DNA (blue). Yellow arrow indicates a centromere. (B) Distributions of transfected GFP-Brd4 and deletions were analyzed on mitotic chromosomes prepared from cells treated with or without TSA.
Similar articles
- Structural basis and binding properties of the second bromodomain of Brd4 with acetylated histone tails.
Liu Y, Wang X, Zhang J, Huang H, Ding B, Wu J, Shi Y. Liu Y, et al. Biochemistry. 2008 Jun 17;47(24):6403-17. doi: 10.1021/bi8001659. Epub 2008 May 24. Biochemistry. 2008. PMID: 18500820 - Selective recognition of acetylated histones by bromodomains in transcriptional co-activators.
Hassan AH, Awad S, Al-Natour Z, Othman S, Mustafa F, Rizvi TA. Hassan AH, et al. Biochem J. 2007 Feb 15;402(1):125-33. doi: 10.1042/BJ20060907. Biochem J. 2007. PMID: 17049045 Free PMC article. - Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin.
McPhillips MG, Ozato K, McBride AA. McPhillips MG, et al. J Virol. 2005 Jul;79(14):8920-32. doi: 10.1128/JVI.79.14.8920-8932.2005. J Virol. 2005. PMID: 15994786 Free PMC article. - Do protein motifs read the histone code?
de la Cruz X, Lois S, Sánchez-Molina S, Martínez-Balbás MA. de la Cruz X, et al. Bioessays. 2005 Feb;27(2):164-75. doi: 10.1002/bies.20176. Bioessays. 2005. PMID: 15666348 Review. - Biological function and histone recognition of family IV bromodomain-containing proteins.
Lloyd JT, Glass KC. Lloyd JT, et al. J Cell Physiol. 2018 Mar;233(3):1877-1886. doi: 10.1002/jcp.26010. Epub 2017 Jun 13. J Cell Physiol. 2018. PMID: 28500727 Free PMC article. Review.
Cited by
- Lost in transcription: molecular mechanisms that control HIV latency.
Taube R, Peterlin M. Taube R, et al. Viruses. 2013 Mar 21;5(3):902-27. doi: 10.3390/v5030902. Viruses. 2013. PMID: 23518577 Free PMC article. Review. - BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses.
Belkina AC, Nikolajczyk BS, Denis GV. Belkina AC, et al. J Immunol. 2013 Apr 1;190(7):3670-8. doi: 10.4049/jimmunol.1202838. Epub 2013 Feb 18. J Immunol. 2013. PMID: 23420887 Free PMC article. - Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting.
Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Wu SY, et al. Mol Cell. 2013 Mar 7;49(5):843-57. doi: 10.1016/j.molcel.2012.12.006. Epub 2013 Jan 11. Mol Cell. 2013. PMID: 23317504 Free PMC article. - Heterochromatin protein 1 gamma and IκB kinase alpha interdependence during tumour necrosis factor gene transcription elongation in activated macrophages.
Thorne JL, Ouboussad L, Lefevre PF. Thorne JL, et al. Nucleic Acids Res. 2012 Sep;40(16):7676-89. doi: 10.1093/nar/gks509. Epub 2012 May 30. Nucleic Acids Res. 2012. PMID: 22649058 Free PMC article. - BET Bromodomain Inhibition Potentiates Ocular Melanoma Therapy by Inducing Cell Cycle Arrest.
Chen X, Huang R, Zhang Z, Song X, Shen J, Wu Q. Chen X, et al. Invest Ophthalmol Vis Sci. 2024 Jul 1;65(8):11. doi: 10.1167/iovs.65.8.11. Invest Ophthalmol Vis Sci. 2024. PMID: 38967943 Free PMC article.
References
- Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. - PubMed
- Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. - PubMed
- Jeanmougin, F., Wurtz, J. M., Le Douarin, B., Chambon, P. & Losson, R. (1997) Trends Biochem. Sci. 22, 151–153. - PubMed
- Dhalluin, C., Carlson, J. E., Zeng, L., He, C., Aggarwal, A. K. & Zhou, M. M. (1999) Nature 399, 491–496. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases