The Sox9 transcription factor determines glial fate choice in the developing spinal cord - PubMed (original) (raw)
The Sox9 transcription factor determines glial fate choice in the developing spinal cord
C Claus Stolt et al. Genes Dev. 2003.
Abstract
The mechanism that causes neural stem cells in the central nervous system to switch from neurogenesis to gliogenesis is poorly understood. Here we analyzed spinal cord development of mice in which the transcription factor Sox9 was specifically ablated from neural stem cells by the CRE/loxP recombination system. These mice exhibit defects in the specification of oligodendrocytes and astrocytes, the two main types of glial cells in the central nervous system. Accompanying an early dramatic reduction in progenitors of the myelin-forming oligodendrocytes, there was a transient increase in motoneurons. Oligodendrocyte progenitor numbers recovered at later stages of development, probably owing to compensatory actions of the related Sox10 and Sox8, both of which overlap with Sox9 in the oligodendrocyte lineage. In agreement, compound loss of Sox9 and Sox10 led to a further decrease in oligodendrocyte progenitors. Astrocyte numbers were also severely reduced in the absence of Sox9 and did not recover at later stages of spinal cord development. Taking the common origin of motoneurons and oligodendrocytes as well as V2 interneurons and some astrocytes into account, stem cells apparently fail to switch from neurogenesis to gliogenesis in at least two domains of the ventricular zone, indicating that Sox9 is a major molecular component of the neuron-glia switch in the developing spinal cord.
Figures
Figure 1.
Sox9 expression pattern in the embryonic spial cord. Immunohistochemistry was performed on transverse sections from the forelimb region of wild-type embryos with an antibody specifically directed against Sox9. (a) 8.5 dpc. (b) 9.5 dpc. (c) 10.5 dpc. (d) 11.5 dpc. (e) 12.5 dpc. (f) 14.5 dpc. (g) 16.5 dpc. (h) 18.5 dpc.
Figure 2.
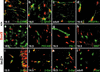
Sox9-expressing cell types in the central nervous system. Immunohistological analysis of embryonic and adult spinal cord (stages as mentioned in the panels) of wild-type (+/+; a_–_h) and Sox10lacZ/+ (lacZ/+; i_–_l) mice using antibodies against Sox9 (red) in combination with various cell-type specific antibodies (green), including B-FABP (a), GFAP (b,c), PDGF receptor α (d), NeuN (e), PECAM-1 (f), Flk-1 (g), MBP (h), and β-galactosidaseSox10 (i_–_l).
Figure 3.
Ablation efficiency and development in Sox9-deficient spinal cords. (a_–_d) Immunohistochemistry with a Sox9-specific antibody on transverse sections from the forelimb region of Sox9Δ/Δ embryos. (a) 10.5 dpc. (b) 12.5 dpc. (c) 14.5 dpc. (d) 16.5 dpc. Note normal Sox9 expression in mesenchymal condensations. (e_–_j) TUNEL labeling of transverse sections from the forelimb region of wild-type (+/+; e_–_g) and Sox9Δ/Δ (h_–_j) spinal cords at 16.5 dpc (e,h), 17.5 dpc (f,i), and 18.5 dpc (g,j). Tissues were counterstained with DAPI (blue). (k) Quantification of TUNEL labeling in wild-type (open bars) and Sox9-deficient (filled bars) spinal cords at 16.5, 17.5, and 18.5 dpc. The number of TUNEL-positive cells obtained for the wild type was arbitrarily set to 1. All other values were expressed relative to wild-type levels and are presented as mean ± S.E.M. (for method, see Figs. 5,7,9). Statistical significance was determined by the Student's t test and is indicated above the respective bars (three asterisks for P ≤ 0.001).
Figure 4.
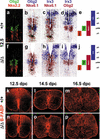
Stem cell and radial glia development in embryonic Sox9-deficient spinal cords. (a_–_j) Immunohistochemistry with antibodies specific for Nkx2.2 and Olig2 (a,f) and in situ hybridizations with antisense probes specific for Olig2, Nkx6.1, and Irx3 (b_–_d,g_–_i) were performed on transverse sections from the forelimb region of embryos at 12.5 dpc to determine the integrity of the three ventral stem cell domains of the ventricular zone (p2, pMN, p3) as summarized in e and j. In situ hybridization signals obtained with different probes on immediately adjacent sections were superimposed using Adobe Photoshop with the color of one of the signals being converted to red. (k_–_p) Immunohistochemistry with antibodies specific for the radial glia marker B-FABP were performed on transverse sections from the forelimb region of embryos. (a_–_e,k_–_m) Wild-type (+/+) spinal cords. (f_–_j,n_–_p) Sox9Δ/Δ spinal cords. (a_–_k,n) 12.5 dpc. (l,o) 14.5 dpc. (m,p) 16.5 dpc.
Figure 5.
Oligodendrocyte development in embryonic Sox9-deficient and Sox9/Sox10-deficient spinal cords. Immunohistochemistry with antibodies specific for the pMN stem cell and oligodendrocyte marker Olig2 (a_–_f), and the oligodendrocyte markers Sox10 (g_–_l) and β-galactosidaseSox10 (m,n) were performed on transverse sections from the forelimb region of embryos. (a_–_c,g_–_i) Wild-type spinal cords. (d_–_f,j_–_l) Sox9Δ/Δ spinal cords. (m,n) Sox9Δ/Δ, Sox10lacZ/lacZ spinal cords. (a,d,g,j,m) 12.5 dpc. (b,e,h,k,n) 14.5 dpc. (c,f,i,l) 16.5 dpc. (o,p) Quantification of oligodendrocytic cells in wild-type (+/+; open bars), Sox9-deficient (Δ/Δ; filled bars), and Sox9/Sox10-deficient (Δ/Δ,–/–; filled bars) spinal cords, as indicated_below_ the bars. The number of Olig2-positive cells (o) or Sox10/β-galactosidaseSox10-positive cells (p) was determined at 12.5 dpc, 14.5 dpc, and 16.5 dpc as indicated. For 12.5 dpc, Olig2-positive cells within (VZ) and outside (MZ) the ventricular zone were separately quantified. At least 15 separate 10-μm sections from the forelimb region of ≥2 independent embryos were counted for each genotype. The number of cells obtained for the wild type was arbitrarily set to 100%. All other values were expressed relative to wild-type levels and are presented as mean ± S.E.M. Statistical significance was determined by the Student's t test and is indicated above the respective bars (three asterisks for P ≤ 0.001).
Figure 6.
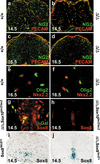
Marker gene expression in Sox9-deficient and Sox9/Sox10-deficient spinal cords. (a_–_d) Immunohistochemistry with antibodies specific for NG2 (green) and PECAM-1 (red) were performed on transverse sections of wild-type (a,c) and Sox9Δ/Δ (b,d) spinal cords from the forelimb region of embryos at 14.5 dpc (a,b) and 16.5 dpc (c,d). The green signal is specific for oligodendrocytes, whereas the yellow color indicates colocalization of NG2 and PECAM-1 in blood vessels (see also Liu et al. 2002). (e,f) Immunohistochemical analysis with antibodies directed against Nkx2.2 (red) and Olig2 (green) on wild-type (e) and Sox9Δ/Δ (f) spinal cords at 16.5 dpc. (g,h) Immunohistochemistry with antibodies directed against Sox class E proteins (red) on Sox9Δ/Δ, Sox10lacZ/lacZ spinal cords at 14.5 dpc, in combination with antibodies against β-galactosidaseSox10 (g; green) or alone (h). (i,j) Comparison of Sox8 expression in Sox8+/lacZ (i) and Sox9Δ/Δ, Sox8+/lacZ (j) spinal cords at 14.5 dpc by X-gal staining for β-galactosidaseSox8.
Figure 7.
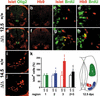
Motoneuron development in embryonic Sox9-deficient spinal cords. Immunohistochemistry with antibodies directed against Islet-1/2 (a,c,e,g,i,j) and Hb9 (b,d,f,h; both in red) were performed on transverse sections from the forelimb region of the embryos, alone (b,f,i,j) or in combination with antibodies directed against Olig2 (a,e) and BrdU (c,d,g,h; both in green) at 12.5 dpc (a_–_h) and 14.5 dpc (i,j). (a_–_d,i) Wild-type spinal cords. (e_–_h,j) Sox9Δ/Δ spinal cords. (k) Quantification of motoneurons in wild-type (open bars) and Sox9-deficient (filled bars) spinal cords. The number of Islet-positive cells at 12.5 dpc was separately determined for the dorsal (region 1, corresponding to D2 interneurons) and the ventral (region 2 + 3, corresponding to motoneurons) spinal cord. Motoneurons were additionally subdivided according to their position (region 2, ventral horn; region 3, medial). At least 35 separate 10-μm sections from the forelimb region of ≥2 independent embryos were counted for each genotype. The number of cells obtained for the wild type was arbitrarily set to 100%. All other values were expressed relative to wild-type levels and are presented as mean ± S.E.M. Statistical significance was determined by the Student's t test and is indicated above the respective bars (two asterisks for P ≤ 0.01, three asterisks for P ≤ 0.001).
Figure 8.
Astrocyte development in embryonic Sox9-deficient spinal cords. Immunohistochemistry with antibodies specific for the astrocyte marker S100β (a,b,d,e) and in situ hybridization with antisense probes specific for the astrocyte markers Glast (c,f), Fgfr3 (g,h,j,k), and glutamine synthase (GlnS; i,l) were performed on transverse sections from the forelimb region of embryos. (a_–_c,g_–_i) Wild-type (+/+) spinal cords. (d_–_f,j_–_l) Sox9Δ/Δ spinal cords. (a,d,g,j) 14.5 dpc. (b,c,e,f,h,i,k,l) 16.5 dpc. (Insets) Costaining of S100β (red) with Chx10 (green;a) and S100β-positive (b) astrocytes at high magnification.
Figure 9.
V2 interneuron development in embryonic Sox9-deficient spinal cords. Immunohistochemistry with antibodies directed against the V2 interneuron marker Chx10 were performed on transverse sections from the forelimb region of embryos. (a_–_c) Wild-type spinal cords. (d_–_f) Sox9Δ/Δ spinal cords. (a,d) 12.5 dpc. (b,e) 14.5 dpc. (c,f) 16.5 dpc. (g) Quantification of Chx10-positive V2 interneurons in wild-type (open bars) and Sox9-deficient (filled bars) spinal cords. The number of Chx10-positive cells was quantified at 12.5, 14.5, and 16.5 dpc as indicated_below_ the bars. At least 35 separate 10-μm sections from the forelimb region of ≥2 independent embryos were counted for each genotype. The number of cells obtained for the wild type was arbitrarily set to 100%. All other values were expressed relative to wild-type levels and are presented as mean ± S.E.M. Statistical significance was determined by the Student's t test and is indicated above the respective bars (three asterisks for P ≤ 0.001).
Figure 10.
Mechanism of Sox9 function in oligodendrocytes. (a,b) RT–PCR analyses on cDNA obtained from Neuro2A cells transiently transfected with >90% efficiency with expression plasmids for GFP (control) or Sox9. Cells were kept under standard culture conditions (a,–RA in_b_) or in retinoic-acid-containing medium to induce neuronal differentiation (+RA in b). Transcript levels of glial markers (Fgfr3, GFAP, GlnS, Sox10, CNP, PLP in a), neuronal markers (AMPA receptor, calbindin D28 in b), and a housekeeping gene (actin) were compared semiquantitatively using increasing numbers of amplification cycles (n, n + 3, n + 6). C, water control. (c) Summary of Sox9 function in the p2 and pMN domains of the embryonic spinal cord. Instead of switching from generation of neurons (V2 interneurons and motoneurons, respectively) to glia (astrocytes and oligodendrocytes, respectively), stem cells in both domains continue to primarily produce neurons of the correct subtype. (d) Summary of Sox gene expression in the oligodendrocyte lineage. Sox9 expression precedes the expression of other class E Sox genes, but is no longer expressed in terminally differentiated oligodendrocytes in contrast to Sox10 and Sox8. This pattern of expression explains the observed oligodendrocyte defects in both Sox9- and Sox10-deficient spinal cords.
Similar articles
- Impact of transcription factor Sox8 on oligodendrocyte specification in the mouse embryonic spinal cord.
Stolt CC, Schmitt S, Lommes P, Sock E, Wegner M. Stolt CC, et al. Dev Biol. 2005 May 15;281(2):309-17. doi: 10.1016/j.ydbio.2005.03.010. Dev Biol. 2005. PMID: 15893981 - Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression.
Finzsch M, Stolt CC, Lommes P, Wegner M. Finzsch M, et al. Development. 2008 Feb;135(4):637-46. doi: 10.1242/dev.010454. Epub 2008 Jan 9. Development. 2008. PMID: 18184726 - Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy.
Stolt CC, Lommes P, Friedrich RP, Wegner M. Stolt CC, et al. Development. 2004 May;131(10):2349-58. doi: 10.1242/dev.01114. Epub 2004 Apr 21. Development. 2004. PMID: 15102707 - Regulation of oligodendrocyte development.
Kagawa T, Wada T, Ikenaka K. Kagawa T, et al. Microsc Res Tech. 2001 Mar 15;52(6):740-5. doi: 10.1002/jemt.1058. Microsc Res Tech. 2001. PMID: 11276126 Review. - Cell fate control in the developing central nervous system.
Guérout N, Li X, Barnabé-Heider F. Guérout N, et al. Exp Cell Res. 2014 Feb 1;321(1):77-83. doi: 10.1016/j.yexcr.2013.10.003. Epub 2013 Oct 16. Exp Cell Res. 2014. PMID: 24140262 Review.
Cited by
- Ascl1/Mash1 promotes brain oligodendrogenesis during myelination and remyelination.
Nakatani H, Martin E, Hassani H, Clavairoly A, Maire CL, Viadieu A, Kerninon C, Delmasure A, Frah M, Weber M, Nakafuku M, Zalc B, Thomas JL, Guillemot F, Nait-Oumesmar B, Parras C. Nakatani H, et al. J Neurosci. 2013 Jun 5;33(23):9752-9768. doi: 10.1523/JNEUROSCI.0805-13.2013. J Neurosci. 2013. PMID: 23739972 Free PMC article. - Context-dependent regulation of Notch signaling in glial development and tumorigenesis.
Guo R, Han D, Song X, Gao Y, Li Z, Li X, Yang Z, Xu Z. Guo R, et al. Sci Adv. 2023 Nov 10;9(45):eadi2167. doi: 10.1126/sciadv.adi2167. Epub 2023 Nov 10. Sci Adv. 2023. PMID: 37948517 Free PMC article. - Induction of specific neuron types by overexpression of single transcription factors.
Teratani-Ota Y, Yamamizu K, Piao Y, Sharova L, Amano M, Yu H, Schlessinger D, Ko MS, Sharov AA. Teratani-Ota Y, et al. In Vitro Cell Dev Biol Anim. 2016 Oct;52(9):961-973. doi: 10.1007/s11626-016-0056-7. Epub 2016 Jun 1. In Vitro Cell Dev Biol Anim. 2016. PMID: 27251161 Free PMC article. - Developmental origins of astrocyte heterogeneity: the final frontier of CNS development.
Chaboub LS, Deneen B. Chaboub LS, et al. Dev Neurosci. 2012;34(5):379-88. doi: 10.1159/000343723. Epub 2012 Nov 9. Dev Neurosci. 2012. PMID: 23147551 Free PMC article. Review. - A critical cell-intrinsic role for serum response factor in glial specification in the CNS.
Lu PP, Ramanan N. Lu PP, et al. J Neurosci. 2012 Jun 6;32(23):8012-23. doi: 10.1523/JNEUROSCI.5633-11.2012. J Neurosci. 2012. PMID: 22674276 Free PMC article.
References
- Bi W., Deng, J.M., Zhang, Z., Behringer, R.R., and de Crombrugghe, B. 1999. Sox9 is required for cartilage formation. Nat. Genet. 22: 85–89. - PubMed
- Bignami A. and Dahl, D. 1974. Astrocyte-specific protein and radial glia in the cerebral cortex of newborn rat. Nature 252: 55–56. - PubMed
- Bowles J., Schepers, G., and Koopman, P. 2000. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227: 239–255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials