Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan - PubMed (original) (raw)
Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan
Andres F Muro et al. J Cell Biol. 2003.
Abstract
Fibronectins (FNs) are multifunctional high molecular weight glycoproteins present in the blood plasma and in the ECMs of tissues. The FN primary transcript undergoes alternative splicing in three regions generating up to 20 main different variants in humans. However, the precise role of the FN isoforms is poorly understood. One of the alternatively spliced exons is the extra domain A (EDA) or extra type III homology that is regulated spatially and temporally during development and aging. To study its in vivo function, we generated mice devoid of EDA exon-regulated splicing. Constitutive exon inclusion was obtained by optimizing the splice sites, whereas complete exclusion was obtained after in vivo CRE-loxP-mediated deletion of the exon. Homozygous mouse strains with complete exclusion or inclusion of the EDA exon were viable and developed normally, indicating that the alternative splicing at the EDA exon is not necessary during embryonic development. Conversely, mice without the EDA exon in the FN protein displayed abnormal skin wound healing, whereas mice having constitutive inclusion of the EDA exon showed a major decrease in the FN levels in all tissues. Moreover, both mutant mouse strains have a significantly shorter lifespan than the control mice, suggesting that EDA splicing regulation is necessary for efficient long-term maintenance of biological functions.
Figures
Figure 1.
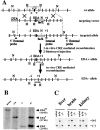
Strategy for the generation of mouse strains lacking regulated splicing at the EDA exon of the FN gene. (A) Partial maps of the wild-type FN allele, the targeting vector, the targeted allele, the EDA-floxed allele, and the EDA-null alleles. Exons, Neo-TK, and DTA are shown as gray, white, and dotted boxes, respectively. The EDA exon and the “loxP” sites are indicated as a black square and as white triangles, respectively. N and H indicate NcoI and HindIII sites, respectively. The recombinant ES cells were transiently transfected with a CRE-recombinase expressing plasmid, and EDA+/wt heterozygote cells were used for blastocyst microinjection. EDA+/wt mice were mated with a CRE-expressing transgenic mouse strain to obtain the EDA−/wt mice. The asterisks indicate the NdeI sites. The 4.0-, 2.7-, and 2.0-kb DNA fragments (EDAwt, EDA+, and EDA− alleles, respectively) obtained after digestion with HindIII and Southern blot hybridization with the internal probe are indicated. (B) Southern blot screening of the different mouse genotypes. EDAwt, EDA+, and EDA− bands are indicated. (C) RT-PCR analysis of total RNA prepared from EDAwt/wt, EDA+/+, and EDA−/− from liver, brain, and kidney is shown. EDA+ and EDA− bands are indicated (805 and 535 bp, respectively).
Figure 2.
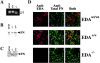
Regulated splicing of the EDA is dispensable for the ECM formation. (A) RT-PCR analysis of total RNA prepared from MEF from EDAwt/wt, EDA+/+, and EDA−/− embryos (13.5 d postcoitus [p.c.]). (B and C) Western blot analysis of protein extracts prepared from the above-described MEF (20 and 100 μg for B and C, respectively) with anti-FN and anti-EDA antibodies, respectively. Coomassie blue staining showed equal loading. (D) MEF prepared from EDAwt/wt, EDA+/+, and EDA−/− embryos (13.5 d p.c.) were plated, fixed, and incubated with anti-EDA antibody (left column) or with anti–total FN antibody (middle column). The merged image is shown in the right column.
Figure 3.
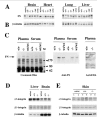
The decrease in FN levels in tissues of EDA +/+ mice is not correlated to a reduction in integrin levels. (A and B) Western blot analysis of total protein extracts (50 μg) from brain, heart, lung, and liver from all genotypes (EDAwt/wt, EDA+/+, EDA−/−, EDA+/−, EDA−/wt, and EDA+/wt) using anti-FN polyclonal antibody (A) and, after stripping the membrane, anti–β-tubulin mAb to verify for minor errors in protein load (B). Note that the migration of the EDA+ FN is slightly slower than that of the EDA− FN. Coomassie blue staining of 5–17% gradient gels of the same protein extracts did not show changes in other proteins. (C) Plasma and serum protein samples from EDA+/+, EDA−/−, and EDAwt/wt mice (20 μg) were Coomassie blue stained (left), and a similar gel was blotted and incubated with anti-FN antibody (middle). For the anti-EDA antibody (right), 100 μg of protein extract were loaded. (D and E) Western blot analysis of α9- and β1-integrin levels in different tissues (liver, brain, and skin) using anti-α9 and -β1 rabbit polyclonal antibodies. Anti–β-tubulin mAb was used to verify for minor errors in protein load.
Figure 4.
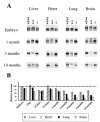
Expression of FN in different tissues at different ages. (A) Protein extracts from liver, heart, lung, and brain were prepared from EDAwt/wt, EDA+/+, and EDA−/− mice of different ages (13.5 d p.c., 1, 3, and 14 mo old) and analyzed by Western blot using an anti-FN antibody. (B) The histograms represent the relative amount of FN present in EDA+/+ mice relative to that present in EDAwt/wt mice (considered as 100%) for each tissue and each time point.
Figure 5.
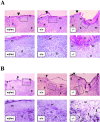
Reepithelization is observed 5 d after full thickness cutaneous wounding. Full thickness cutaneous wounds of control (EDAwt/wt, n = 6) and mutant mice (EDA+/+ and EDA−/−, n = 6 and 7, respectively) mice were analyzed at 5 d after wounding by hematoxylin-eosin staining. Representative sections are shown. The black arrowheads indicate the wound edges. Epithelium, granulation tissue, dermis, newly formed epithelium, and eschar are indicated (“e,” “g,” “d,” “re”, and “sc,” respectively). The dotted rectangles in the top panels indicate the magnified area showed in the bottom panels. The experiment was repeated twice with similar results. Edematous granulation tissue is observed in the EDA−/− wounds.
Figure 6.
EDA − / − skin wounds at day 7 after wounding show abnormal healing and increased number of BrdU-positive cells. (A) Full thickness cutaneous wounds of control (EDAwt/wt, n = 8) and mutant mice (EDA+/+ and EDA−/−, n = 8 and 9, respectively) mice were analyzed at 7 d after wounding. Representative sections are shown. The black arrowheads indicate the wound edges. Abbreviations are as described in Fig. 5. The ulcerated epithelium observed in the EDA−/− wound is indicated as “ue.” The experiment was repeated three times with similar results. Wound sections of three independent experiments (eight mice per genotype in each experiments) were microscopically scored for the presence of ulcerative processes at day 7 after wounding. The fraction of ulcerated wounds of the EDA−/− mice was statistically significant different from that of EDAwt/wt and EDA+/+ mice (63 and 22% for EDA−/− and EDAwt/wt, respectively; P < 0.0002 for EDA−/− vs. EDAwt/wt by both the Fisher Exact test and the χ2 test). There were no differences between the EDAwt/wt and EDA+/+ scores). (B) BrdU labeling shows an increased number of replicating cells in the EDA−/− wounds. Serial sections of the same wounds seen in A were incubated with an anti-BrdU mAb. The count of several microscopical fields showed that there are at least 10 times more BrdU-positive nuclei in the area below the newly formed epidermis in the EDA−/− wounds when compared with EDAwt/wt or EDA+/+ wounds. A representative field is shown. More than 50 positive nuclei are observed in the EDA−/− wound (bottom right), whereas less than five positive nuclei are observed in the case of EDAwt/wt or EDA+/+ samples (bottom left and center, respectively). Illustrative BrdU-labeled nuclei are indicated by white triangles. The black arrows indicate the wound edges.
Figure 7.
The lack of alternative splicing of the EDA exon reduces the lifespan of mutant mice. Both EDA+/+ and EDA−/− mice showed a reduced lifespan during the 30-mo period of analysis. The graphic shows a Kaplan-Meier representation of the survival versus time of 39, 45, and 53 EDAwt/wt, EDA+/+, and EDA−/− mice, respectively, that were housed in individual cages throughout the study. The results were analyzed with the Log-rank test and both the EDA+/+ and the EDA−/− curves were statistically significant when compared with the EDAwt/wt mice (indicated by asterisks, P ≤ 0.0005).
Similar articles
- Impaired motor coordination in mice lacking the EDA exon of the fibronectin gene.
Chauhan AK, Moretti FA, Iaconcig A, Baralle FE, Muro AF. Chauhan AK, et al. Behav Brain Res. 2005 Jun 3;161(1):31-8. doi: 10.1016/j.bbr.2005.02.020. Epub 2005 Apr 12. Behav Brain Res. 2005. PMID: 15904707 - Age-dependent differential expression of fibronectin variants in skin and airway mucosal wounds.
Li-Korotky HS, Hebda PA, Lo CY, Dohar JE. Li-Korotky HS, et al. Arch Otolaryngol Head Neck Surg. 2007 Sep;133(9):919-24. doi: 10.1001/archotol.133.9.919. Arch Otolaryngol Head Neck Surg. 2007. PMID: 17875859 - Alternatively spliced isoforms of fibronectin in immune-mediated glomerulosclerosis: the role of TGFbeta and IL-4.
Baelde HJ, Eikmans M, van Vliet AI, Bergijk EC, de Heer E, Bruijn JA. Baelde HJ, et al. J Pathol. 2004 Nov;204(3):248-57. doi: 10.1002/path.1653. J Pathol. 2004. PMID: 15372454 - Alternative splicing of fibronectin: three variants, three functions.
Schwarzbauer JE. Schwarzbauer JE. Bioessays. 1991 Oct;13(10):527-33. doi: 10.1002/bies.950131006. Bioessays. 1991. PMID: 1755828 Review. - Fibronectin expression in the cardiovascular system.
Sabri A, Farhadian F, Contard F, Samuel JL, Rappaport L. Sabri A, et al. Herz. 1995 Apr;20(2):118-26. Herz. 1995. PMID: 7774863 Review.
Cited by
- Fibronectin in tissue regeneration: timely disassembly of the scaffold is necessary to complete the build.
Stoffels JM, Zhao C, Baron W. Stoffels JM, et al. Cell Mol Life Sci. 2013 Nov;70(22):4243-53. doi: 10.1007/s00018-013-1350-0. Epub 2013 Jun 12. Cell Mol Life Sci. 2013. PMID: 23756580 Free PMC article. Review. - Substance P induces fibrotic changes through activation of the RhoA/ROCK pathway in an in vitro human corneal fibrosis model.
Słoniecka M, Danielson P. Słoniecka M, et al. J Mol Med (Berl). 2019 Oct;97(10):1477-1489. doi: 10.1007/s00109-019-01827-4. Epub 2019 Aug 9. J Mol Med (Berl). 2019. PMID: 31399750 Free PMC article. - Direct test of potential roles of EIIIA and EIIIB alternatively spliced segments of fibronectin in physiological and tumor angiogenesis.
Astrof S, Crowley D, George EL, Fukuda T, Sekiguchi K, Hanahan D, Hynes RO. Astrof S, et al. Mol Cell Biol. 2004 Oct;24(19):8662-70. doi: 10.1128/MCB.24.19.8662-8670.2004. Mol Cell Biol. 2004. PMID: 15367684 Free PMC article. - Assembly of fibronectin extracellular matrix.
Singh P, Carraher C, Schwarzbauer JE. Singh P, et al. Annu Rev Cell Dev Biol. 2010;26:397-419. doi: 10.1146/annurev-cellbio-100109-104020. Annu Rev Cell Dev Biol. 2010. PMID: 20690820 Free PMC article. Review. - Smooth muscle cell-specific fibronectin-EDA mediates phenotypic switching and neointimal hyperplasia.
Jain M, Dhanesha N, Doddapattar P, Chorawala MR, Nayak MK, Cornelissen A, Guo L, Finn AV, Lentz SR, Chauhan AK. Jain M, et al. J Clin Invest. 2020 Jan 2;130(1):295-314. doi: 10.1172/JCI124708. J Clin Invest. 2020. PMID: 31763999 Free PMC article.
References
- Boyle, D.L., Y. Shi, S. Gay, and G.S. Firestein. 2000. Regulation of CS1 fibronectin expression and function by IL-1 in endothelial cells. Cell. Immunol. 200:1–7. - PubMed
- Caputi, M., F.E. Baralle, and C.A. Melo. 1995. Analysis of the linkage between fibronectin alternative spliced sites during ageing in rat tissues. Biochim. Biophys. Acta. 1263:53–59. - PubMed
- Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156–159. - PubMed
- Clark, R.A., J.M. Lanigan, P. DellaPelle, E. Manseau, H.F. Dvorak, and R.B. Colvin. 1982. Fibronectin and fibrin provide a provisional matrix for epidermal cell migration during wound reepithelialization. J. Invest. Dermatol. 79:264–269. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous