Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7 - PubMed (original) (raw)
Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7
Ariberto Fassati et al. EMBO J. 2003.
Abstract
Human immunodeficiency virus type 1 (HIV-1), like other lentiviruses, can infect non-dividing cells. This property depends on the active nuclear import of its intracellular reverse transcription complex (RTC). We have studied nuclear import of purified HIV-1 RTCs in primary macrophages and found that importin 7, an import receptor for ribosomal proteins and histone H1, is involved in the process. Nuclear import of RTCs requires, in addition, energy and the components of the Ran system. Depletion of importin 7 from cultured cells by small interfering RNA inhibits HIV-1 infection. These results provide a new insight into the molecular mechanism for HIV-1 nuclear import and reveal potential targets for therapeutic intervention.
Figures
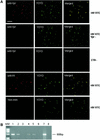
Fig. 1. Analyses of purified HIV-1 RTCs by confocal microscopy and endogenous reverse transcription assay. (A) Purified RTCs were adsorbed onto a plastic tissue culture dish, fixed and labelled with the nucleic acid dye YOYO-1 and anti-Vpr or anti-IN antibodies. Images were acquired sequentially and merged using the Confocal Assistant software. Mutant RTCs lacking Vpr (RTC Vpr–), samples from uninfected cells (CTR–) and non- immune rabbit sera were used as controls. Scale bar, 15 µm. (B) Endogenous RT assay on the equilibrium density fractions containing the peak of the viral DNA. Samples were incubated in the presence or absence of exogenous dNTPs and then subjected to PCR with primers specific for the (+) strand DNA (expected band size is 600 bp). MW, DNA molecular weight markers; 1, RTC fraction incubated in the presence of exogenous dNTPs; 2, same fraction without dNTPs; 3, fraction from uninfected cells; 4–6, same conditions as in 1–3 but RTCs were extracted in 160 mM KCl buffer after the first extraction in hypotonic buffer; 7, positive control; 8, no DNA.
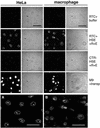
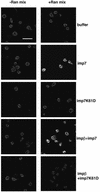
Fig. 2. Nuclear import assay with labelled RTCs in HeLa cells and primary human macrophages. Following permeabilization with digitonin, cells were incubated with 103 RTCs/cell (RTC+) or an equal volume of sample from uninfected cells (CTR–) or 1 µM fluorescent-labelled M9-bearing nucleoplasmin core (M9) plus transportin (transp, 1 µM). Incubation was performed for 15 min at 37°C in the presence of energy mix (E), Ran mix (R) and HeLa high-speed cytosolic exctracts (HSE, 0.5 mg/ml final concentration) or buffer. Fluorescent and transmission images of the same fields are shown. Scale bars, 25 µm. (A) Enlarged image corresponding to the HeLa RTC HSE panel. (B) Enlarged image corresponding to the macrophage RTC HSE panel. Scale bars, 25 µm.
Fig. 3. Time-lapse microscopy of RTC nuclear import in permeabilized HeLa cells. (A) Permeabilized cells were placed on a heated plate (37°C) and incubated in the presence of 0.5 mg/ml HeLa HSE, 103 labelled RTCs/cell, 1× energy mix and Ran mix. Images were acquired every 30 s using a Zeiss Axiovert S100 fluorescent microscope over a 25 min period, and images were processed using the Meta Imaging series 4.5 (Universal Imaging Corp.). Arrows point to fluorescent nucleoli; scale bar, 20 µm. (B) PCR analysis of viral DNA in HeLa cells after the nuclear import assay with purified RTCs. Following large-scale nuclear import assays, Hirt DNA was extracted (Hirt, 1967) and analysed by PCR with primers specific for the (+) strand DNA, a late reverse transcription product. 1, cells incubated in the presence of HSE, Ran mix and energy mix; 2, cells incubated in the absence of HSE; 3, cells incubated with the fraction from uninfected cells; 4, cells infected with the HIV-1 vector; 5, uninfected cells.
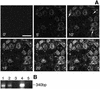
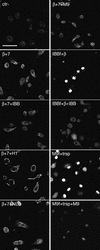
Fig. 4. Imp7 and the impβ–imp7 heterodimer stimulate nuclear import of RTCs. Primary human macrophages were permeabilized with digitonin and incubated at 37°C for 15 min in the presence of 103 labelled RTCs/cell, 1× energy mix, 1× Ran mix and the following import receptors: 0.5 µM each impα + impβ (α+β), 0.5 µM impβ (β), 0.5 µM imp4, imp5, imp7, imp9A, imp13, transportin (transp) and 0.5 µM imp7 + 0.25 µM impβ (imp7+β). MBP fused to the M9 peptide (M9) and the ribosomal protein L23a (RpL23a) were used as positive controls at 1 µM each + 1 µM specific import receptor. Cells were fixed and analysed by confocal microscopy. Scale bar, 30 µm.
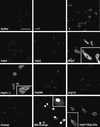
Fig. 5. Histone H1 inhibits RTC nuclear import in primary macrophages. Primary human macrophages were permeabilized with digitonin and incubated at 37°C for 15 min with 103 labelled RTCs/cell, 1× energy mix, 0.75× Ran mix, 0.75 µM each impβ and imp7 (β+7) or without importins (Ctr–) and 1.5 µM of the following recombinant factors as indicated: MBP–IBB (IBB), MBP–M9 (M9), histone H1 (H1) and BSA–NLS (NLS). Control samples were incubated in the same conditions with 0.5 µM fluorescently labelled GST–IBB (IBBf) + 1 µM impβ or labelled MBP–M9 (M9f) + 1 µM transportin with or without their specific competitors (IBB and M9, respectively). Cells were fixed and analysed by confocal microscopy. Scale bar, 25 µm.
Fig. 6. Effect of the Ran system and the mutant imp7 K61D on nuclear import of RTCs. Primary human macrophages were treated as in Figure 3 and incubated at 37°C for 15 min with 103 labelled RTCs/cell, 1 µM import receptors with or without energy and Ran mixes. The mutant imp7 K61D is unable to bind Ran. Samples were fixed and analysed by confocal microscopy. Scale bar, 25 µm.
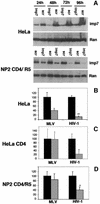
Fig. 7. Imp7 depletion inhibits HIV-1 infection. (A) Western blot showing specific depletion of imp7 by siRNA in HeLa cells and NP2 cells expressing CD4 and CCR5. Cells were transfected with 60 pmol siRNA duplex and analysed by western blot 24–96 h post-transfection. Imp7, anti-importin 7 siRNA; scr, scramble siRNA. Levels of Ran are shown as loading controls. (B) HeLa cells transfected with imp7 siRNA (grey bars) or with a scramble siRNA (black bars) were infected at an m.o.i. of 0.01 with HIV-1 or MLV vectors pseudotyped with VSV-G. (C) HeLa CD4 cells transfected with imp7 siRNA (grey bars) or scramble siRNA (black bars) were infected with the HIV-1 vector harbouring the gp120 envelope or an MLV vector pseudotyped with VSV-G. (D) NP2 cells, similarly transfected with imp7 siRNA (grey bars) or scramble siRNA (black bars), were infected at the same m.o.i. with SF-162 HIV-1 primary isolate or with amphotropic MLV. Infected cells were counted 48 h post-infection. Values are expressed as mean number of infected cells ± SD. HIV titres in imp7-depleted cells were reduced compared with both scramble siRNA-treated cells and MLV-infected cells (paired Student _t_-test, P < 0.004).
Similar articles
- tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes.
Zaitseva L, Myers R, Fassati A. Zaitseva L, et al. PLoS Biol. 2006 Oct;4(10):e332. doi: 10.1371/journal.pbio.0040332. PLoS Biol. 2006. PMID: 17020411 Free PMC article. - HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome.
Zaitseva L, Cherepanov P, Leyens L, Wilson SJ, Rasaiyaah J, Fassati A. Zaitseva L, et al. Retrovirology. 2009 Feb 4;6:11. doi: 10.1186/1742-4690-6-11. Retrovirology. 2009. PMID: 19193229 Free PMC article. - Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication.
Ao Z, Huang G, Yao H, Xu Z, Labine M, Cochrane AW, Yao X. Ao Z, et al. J Biol Chem. 2007 May 4;282(18):13456-67. doi: 10.1074/jbc.M610546200. Epub 2007 Mar 14. J Biol Chem. 2007. PMID: 17360709 - HIV-1 nuclear import: in search of a leader.
Bukrinsky MI, Haffar OK. Bukrinsky MI, et al. Front Biosci. 1997 Dec 1;2:d578-87. doi: 10.2741/a213. Front Biosci. 1997. PMID: 9366553 Review. - Nuclear Import of HIV-1.
Shen Q, Wu C, Freniere C, Tripler TN, Xiong Y. Shen Q, et al. Viruses. 2021 Nov 8;13(11):2242. doi: 10.3390/v13112242. Viruses. 2021. PMID: 34835048 Free PMC article. Review.
Cited by
- Interaction of the HIV-1 intasome with transportin 3 protein (TNPO3 or TRN-SR2).
Larue R, Gupta K, Wuensch C, Shkriabai N, Kessl JJ, Danhart E, Feng L, Taltynov O, Christ F, Van Duyne GD, Debyser Z, Foster MP, Kvaratskhelia M. Larue R, et al. J Biol Chem. 2012 Oct 5;287(41):34044-58. doi: 10.1074/jbc.M112.384669. Epub 2012 Aug 7. J Biol Chem. 2012. PMID: 22872640 Free PMC article. - A phenotypic recessive, post-entry block in rabbit cells that results in aberrant trafficking of HIV-1.
Cutiño-Moguel T, Fassati A. Cutiño-Moguel T, et al. Traffic. 2006 Aug;7(8):978-92. doi: 10.1111/j.1600-0854.2006.00449.x. Traffic. 2006. PMID: 16882040 Free PMC article. - Early steps of retrovirus replicative cycle.
Nisole S, Saïb A. Nisole S, et al. Retrovirology. 2004 May 14;1:9. doi: 10.1186/1742-4690-1-9. Retrovirology. 2004. PMID: 15169567 Free PMC article. Review. - Stimulated nuclear import by β-like importins.
Flores K, Seger R. Flores K, et al. F1000Prime Rep. 2013 Oct 1;5:41. doi: 10.12703/P5-41. F1000Prime Rep. 2013. PMID: 24167722 Free PMC article. Review. - Slower uncoating is associated with impaired replicative capability of simian-tropic HIV-1.
Kono K, Takeda E, Tsutsui H, Kuroishi A, Hulme AE, Hope TJ, Nakayama EE, Shioda T. Kono K, et al. PLoS One. 2013 Aug 13;8(8):e72531. doi: 10.1371/journal.pone.0072531. eCollection 2013. PLoS One. 2013. PMID: 23967315 Free PMC article.
References
- Adam S.A. and Gerace,L. (1991) Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell, 66, 837–847. - PubMed
- Bischoff F.R., Scheffzek,K. and Postingl,H. (2002) How Ran is regulated. In Weis,K. (ed.), Nuclear Transport. Springer-Verlag, Berlin, Germany, pp. 49–66.
- Bukrinsky M.I., Sharova,N., McDonald,T.L., Pushkarskaya,T., Tarpley,W.G. and Stevenson M. (1993) Association of integrase, matrix and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl Acad. Sci. USA, 90, 6125–6129. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous