Simultaneously monitoring DNA binding and helicase-catalyzed DNA unwinding by fluorescence polarization - PubMed (original) (raw)
Simultaneously monitoring DNA binding and helicase-catalyzed DNA unwinding by fluorescence polarization
H Q Xu et al. Nucleic Acids Res. 2003.
Abstract
A new method for helicase-catalyzed DNA unwinding is described. This assay takes advantage of the substantial change in fluorescence polarization (FP) upon helicase binding and DNA unwinding. The low anisotropy value, due to the fast tumbling of the free oligonucleotide in solution, increases abruptly upon binding of helicase to the fluorescein-labeled oligonucleotide. The high anisotropy of the helicase- DNA complex decreases as the fluorescein-labeled oligonucleotide is released from the complex through helicase-catalyzed DNA unwinding. This FP signal can be measured in real time by fluorescent spectroscopy. This assay can simultaneously monitor DNA binding and helicase-catalyzed DNA unwinding. It can also be used to determine the polarity in DNA unwinding mediated by helicase. This FP assay should facilitate the study of the mechanism by which helicase unwinds duplex DNA, and also aid in screening for helicase inhibitors, which are of growing interest as potential anticancer agents.
Figures
Figure 1
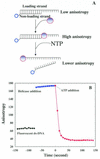
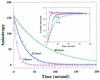
(A) Schematic illustration of the FP-based unwinding assay. A fluorescein-labeled oligonucleotide (open circles) was annealed to the proximal end of a ssDNA molecule. This substrate alone gives a low anisotropy value. The anisotropy signal increases upon the binding of helicase (closed circles) to this DNA substrate. When the DNA strands are unwound by a helicase, the fluorescein-labeled oligo is released from the DNA–helicase complexes, and the anisotropy value is even lower than partial duplex DNA substrate. The unwinding reaction is followed by measurement of the anisotropy value. (B) DNA unwinding followed by fluorescence anisotropy. Fluorescein-labeled partial duplex DNA (5 nM) (black closed circles) was incubated in the unwinding buffer. The anisotropy value was increased and then decreased upon addition of 30 nM RecQ and 1 mM ATP, respectively.
Figure 2
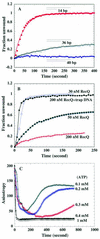
Effect of fluorescein-labeled DNA length protein and ATP concentration on the kinetics of duplex unwinding. (A) Kinetics of duplex unwinding with DNA formed with different lengths of fluorescein-labeled ssDNA as indicated. A 5 nM concentration of each DNA substrate and 40 nM RecQ protein were used in this study. (B) DNA substrate E (2 nM) (36 base duplex DNA) was incubated with 50 and 200 nM RecQ helicase. The DNA unwinding reaction was initiated by adding either 1 mM ATP (closed squares and open circles) or 1 mM ATP containing 2 µM ssDNA trap (closed circles). (C) DNA substrate A (2 nM) was incubated with 40 nM RecQ helicase. The unwinding reaction was initiated by the addition of different concentrations of ATP as indicated.
Figure 3
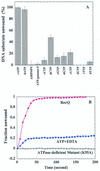
(A) Effects of nucleotides on RecQ helicase-catalyzed DNA unwinding activity. DNA unwinding assays were performed as described in Materials and Methods using DNA substrate A (2 nM); each reaction contained 40 nM helicases and one of the listed nucleotides at a final concentration of 1 mM. The data presented are the average of three independent determinations. (B) Helicase-mediated DNA unwinding activity is ATP dependent. DNA substrate A (5 nM) was incubated with either 30 nM RecQ helicase or 30 nM ATPase-deficient mutant (K55A) enzyme and the unwinding reaction was initiated by addition of either 1 mM ATP (closed circles, wt helicase; open squares, mutant helicase) or 1 mM ATP containing 5 mM EDTA (closed squares).
Figure 4
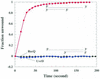
RecQ helicase resides on the loading strand of DNA whereas the leading strand is excluded from the DNA–helicase complexes during the unwinding reaction. DNA substrates A, C, D (5 nM) were incubated with 30 nM RecQ helicase at 25°C and 1 mM ATP was then added. The anisotropy values were recorded every 8 s.
Figure 5
The effect of duplex DNA length on the single-turnover kinetics of RecQ-mediated DNA unwinding. RecQ helicase (50 nM) was incubated with 2 nM DNA substrtate A (circles, 14 bases in duplex length), substrate B (squares, 22 bases in duplex length), substrate E (rhombus, 40 bases in duplex length); DNA unwinding was initiated upon addition of 1 mM ATP containing 2 µM ssDNA trap at 25°C. Data from all three time courses were fitted to the exponential equation: A t = A exp(–k_obs_t), where A t is the anisotropy amplitude at time t, and _k_obs is the observed rate constant. The insert shows the fraction unwound of DNA substrates.
Figure 6
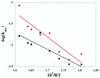
Arrhenius plots of the observed unwinding rate of RecQ helicase. The experiments were performed under standard conditions at temperatures between 4 and 37°C. DNA substrates A and B (2 nM) and 40 nM RecQ helicase were used.
Figure 7
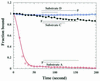
Helicase polarity determination by fluorescence anisotropy assay. RecQ helicase (30 nM) or UvrD helicase (30 nM) was incubated, respectively, with 2 nM partial duplex DNA substrate F or G, and DNA unwinding was initiated by the addition of 1 mM ATP.
Similar articles
- Monitoring helicase-catalyzed DNA unwinding by fluorescence anisotropy and fluorescence cross-correlation spectroscopy.
Xi XG, Deprez E. Xi XG, et al. Methods. 2010 Jul;51(3):289-94. doi: 10.1016/j.ymeth.2010.02.022. Epub 2010 Feb 26. Methods. 2010. PMID: 20219681 Review. - Single-turnover kinetics of helicase-catalyzed DNA unwinding monitored continuously by fluorescence energy transfer.
Bjornson KP, Amaratunga M, Moore KJ, Lohman TM. Bjornson KP, et al. Biochemistry. 1994 Nov 29;33(47):14306-16. doi: 10.1021/bi00251a044. Biochemistry. 1994. PMID: 7947840 - Measurement of steady-state kinetic parameters for DNA unwinding by the bacteriophage T4 Dda helicase: use of peptide nucleic acids to trap single-stranded DNA products of helicase reactions.
Nanduri B, Eoff RL, Tackett AJ, Raney KD. Nanduri B, et al. Nucleic Acids Res. 2001 Jul 1;29(13):2829-35. doi: 10.1093/nar/29.13.2829. Nucleic Acids Res. 2001. PMID: 11433029 Free PMC article. - RecQ helicase-catalyzed DNA unwinding detected by fluorescence resonance energy transfer.
Zhang XD, Dou SX, Xie P, Wang PY, Xi XG. Zhang XD, et al. Acta Biochim Biophys Sin (Shanghai). 2005 Sep;37(9):593-600. doi: 10.1111/j.1745-7270.2005.00084.x. Acta Biochim Biophys Sin (Shanghai). 2005. PMID: 16143813 - Mechanisms of helicase-catalyzed DNA unwinding.
Lohman TM, Bjornson KP. Lohman TM, et al. Annu Rev Biochem. 1996;65:169-214. doi: 10.1146/annurev.bi.65.070196.001125. Annu Rev Biochem. 1996. PMID: 8811178 Review.
Cited by
- Specific activation of the TLR1-TLR2 heterodimer by small-molecule agonists.
Cheng K, Gao M, Godfroy JI, Brown PN, Kastelowitz N, Yin H. Cheng K, et al. Sci Adv. 2015;1(3):e1400139. doi: 10.1126/sciadv.1400139. Sci Adv. 2015. PMID: 26101787 Free PMC article. - Direct observation of the translocation mechanism of transcription termination factor Rho.
Gocheva V, Le Gall A, Boudvillain M, Margeat E, Nollmann M. Gocheva V, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2367-77. doi: 10.1093/nar/gkv085. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662222 Free PMC article. - Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery.
Tuteja N, Tuteja R. Tuteja N, et al. Eur J Biochem. 2004 May;271(10):1835-48. doi: 10.1111/j.1432-1033.2004.04093.x. Eur J Biochem. 2004. PMID: 15128294 Free PMC article. Review. - Establishment and Application of a High Throughput Screening System Targeting the Interaction between HCV Internal Ribosome Entry Site and Human Eukaryotic Translation Initiation Factor 3.
Zhu Y, Huang P, Yang N, Liu R, Liu X, Dai H, Zhang L, Song F, Sun C. Zhu Y, et al. Front Microbiol. 2017 May 29;8:977. doi: 10.3389/fmicb.2017.00977. eCollection 2017. Front Microbiol. 2017. PMID: 28611766 Free PMC article. - Label-free luminescence switch-on detection of hepatitis C virus NS3 helicase activity using a G-quadruplex-selective probe.
Leung KH, He HZ, He B, Zhong HJ, Lin S, Wang YT, Ma DL, Leung CH. Leung KH, et al. Chem Sci. 2015 Apr 1;6(4):2166-2171. doi: 10.1039/c4sc03319a. Epub 2014 Nov 25. Chem Sci. 2015. PMID: 28808523 Free PMC article.
References
- Hall M.C. and Matson,S.W. (1999) Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol., 35, 867–877. - PubMed
- Matson S.W., Bean,D.W. and George,J.W. (1994) DNA helicases enzymes with essential roles in all aspects of DNA metabolism. Bioessays, 16, 13–22. - PubMed
- Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. - PubMed
- Schmid S.R. and Linder,P. (1992) D-E-A-D protein family of putative RNA helicases. Mol. Microbiol., 6, 283–291. - PubMed
- Ellis N.A. (1997) DNA helicases in inherited human disorders. Curr. Opin. Genet. Dev., 7, 354–363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources