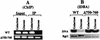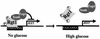Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1 - PubMed (original) (raw)
Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1
Jeong-Ho Kim et al. Mol Cell Biol. 2003 Aug.
Abstract
Rgt1 is a glucose-responsive transcription factor that binds to the promoters of several HXT genes encoding glucose transporters in Saccharomyces cerevisiae and regulates their expression in response to glucose. Rgt1 contains a Zn(2)Cys(6) binuclear cluster responsible for DNA binding. Most proteins that contain this sequence motif bind as dimers to regularly spaced pairs of the sequence CGG. However, there are no CGG pairs with regular spacing in promoters of genes regulated by Rgt1, suggesting that Rgt1 binds as a monomer to CGG or to another sequence. We identified the Rgt1 consensus binding site sequence 5'-CGGANNA-3', multiple copies of which are present in all HXT promoters regulated by Rgt1. Rgt1 binds in vivo to multiple sites in the HXT3 promoter in a nonadditive, synergistic manner, leading to synergistic repression of HXT3 transcription. We show that glucose inhibits the DNA-binding ability of Rgt1, thereby relieving repression of HXT gene expression. This regulation of Rgt1 DNA-binding activity is caused by its glucose-induced phosphorylation: the hyperphosphorylated Rgt1 present in cells growing on high levels of glucose does not bind DNA in vivo or in vitro; dephosphorylation of this form of Rgt1 in vitro restores its DNA-binding ability. Furthermore, an altered Rgt1 that functions as a constitutive repressor remains hypophosphorylated when glucose is added to cells and binds DNA under these conditions. These results suggest that glucose regulates the DNA-binding ability of Rgt1 by inducing its phosphorylation.
Figures
FIG. 1.
Identification of Rgt1 binding sites upstream of HXT genes. (A) Diagram of arrangement of sequences upstream of HXT3, HXT1, and HXT2 that are protected from DNase I digestion and DMS methylation. Arrows, locations and orientations of Rgt1 binding sites. (B) Alignment of the sequences footprinted by Rgt1. The number of bases from the central G residue of the 5′-C
G
G-3′ triplet to the ATG codon are in parentheses. The sequence 5′-CGGANNA-3′ is the Rgt1 consensus binding site.
FIG. 2.
Probing the interaction between Rgt1 and its binding sites. (A) Footprinting assays using DNase I, DMS, and UV as probes were carried out with a DNA fragment of the HXT3 promoter containing five Rgt1 binding sites (−372 to −572 [Fig. 3A]) as described in Materials and Methods. For DNase I and DMS protections, a 32P-labeled DNA fragment (2 × 104 cpm) was incubated with different amounts of Rgt1 (30, 60, and 120 ng, from the third gel lane from the left). The first (L) and second gel lanes are the A+G ladder and the control without Rgt1, respectively. For UV photofootprinting, Rgt1 (100 ng) was incubated (+) with the same amounts of DNA used for DNase I and DMS protections, and then DNA-protein complexes were irradiated with UV for the time indicated above each lane. The Rgt1 binding sites are indicated by boxes. (B) Summary of interactions between Rgt1 and its binding sites observed from three different footprinting assays. DMS protection assays show that Rgt1 weakly contacts CGG at C1 and C2 (open circles).
FIG. 3.
Binding of Rgt1 to the HXT3 promoter. (A) EMSA was carried out with a DNA fragment containing five Rgt1 binding sites. A constant amount of 32P-labeled DNA fragment (3 × 104 cpm) was incubated with increasing amounts of Rgt1 as indicated above the gel lanes. Arrows, locations and orientations of Rgt1 binding sites. Roman numerals denote Rgt1-DNA complexes. (B) The five Rgt1 sites were separated and used as probes for DNase I footprinting assays. After incubation of a 32P-labeled DNA fragment (2 × 105 cpm) with increasing amounts of Rgt1 (1, 3, 6, 10, 30, 60, 100, 300, and 600 ng per lane, from the third gel lane from the left) for 20 min, the DNA-protein complexes were digested with 2 U of DNase I for 1 min. The first and second gel lanes are the A+G ladder and the control (without Rgt1), respectively. Site C consists of two Rgt1 binding sites, C1 and C2, 3 bp apart. The binding affinities of Rgt1 were estimated by comparing the intensities (measured on a PhosphorImager by the ImageQuaNT program) of a reference band (indicated by arrows) and a band within the Rgt1 footprint (indicated by boxes).
FIG. 4.
Multiple Rgt1 binding sites mediate synergistic repression. (A) Three different segments of the HXT3 promoter were fused to a HIS3_-lacZ reporter gene (pBM2832) and introduced into yeast wild-type (WT) (YM4127) and Δ_rgt1 (YM4509) strains. Expression of the reporter gene was measured in cells grown on minimal medium containing 2% galactose (Gal) to mid-log phase and then switched to glucose (4%) (Glu) for 1.5 h. (B) Single or multiple copies of Rgt1 sites in cluster I were inserted in the reporter plasmid (pBM2832) assayed for the function of Rgt1 binding sites as described for panel A. These Rgt1 binding sites were generated by annealing complementary oligonucleotides containing one or two Rgt1 sites or by amplifying more than three Rgt1 sites. (C) Mutations in Rgt1 binding sites were generated by recombinational gap repair. Repression is the ratio of β-galactosidase activity in the WT (YM4127) to that in Δ_rgt1_ (YM4509). Arrows (B and C), locations and orientations of Rgt1 binding sites.
FIG. 5.
Rgt1 is synergistically recruited to multiple binding sites in vivo. (A) The same plasmids used for the repression assay shown in Fig. 4 were used for a ChIP assay. The arrows indicate the locations and orientations of Rgt1 binding sites. (B) Chromatins prepared from cells (YM4127) containing the reporter plasmids grown under repressing conditions were immunoprecipitated with anti-Rgt1 antibody. Rgt1 binding sites in the immunoprecipitated DNA (IP) were PCR amplified using a primer set (OM3128 and OM3129) containing [α-32P]dATP and analyzed in a 6% polyacrylamide gel. (C) The amounts of the input DNA and immunoprecipitated DNA were measured by a PhosphorImager, quantified by the ImageQuaNT program, and presented as the ratio of immunoprecipitated counts to input counts. Fig. 8C, box, shows the control of ChIP using anti-Rgt1 antibody.
FIG. 6.
Rgt1 binds with Ssn6-Tup1 on an HXT promoter. Chromatin prepared from yeast cells (WT, YM4127; Δ_ssn6_, YM4554; Δ_rgt1_, YM4509) grown on YP containing different carbon sources as indicated above each lane was immunoprecipitated using anti-Ssn6 antibody (Santa Cruz). The HXT3 promoter in the immunoprecipitated DNA (IP) was amplified in a PCR, resolved in an agarose gel (2%), and visualized by ethidium bromide staining.
FIG. 7.
Glucose does not regulate Rgt1 levels or nuclear localization. (A) Yeast cells (YM4509) expressing LexA-Rgt1 (expressed from pBM3307) were grown on minimal medium containing either 2% galactose or 2% raffinose (equivalent to low levels of glucose) or 4% glucose. Yeast cells (YM4509) expressing only LexA (pBM2662) were grown on minimal medium containing 2% galactose (Vector). LexA-Rgt1 was immunoprecipitated and subjected to Western blot analysis using anti-LexA antibody as a probe. (B) Yeast cells (YM4127) expressing GFP-Rgt1 (expressed from pBM3911) were grown to mid-log phase under repressing conditions (2% galactose), the carbon source was switched to 4% glucose, and growth was continued for 1.5 h. The cells were harvested and stained with DAPI before and after the carbon source was switched and then were imaged for DAPI and GFP fluorescence or by Nomarski optics.
FIG. 8.
Association of Rgt1 with the HXT promoters was determined by ChIP in cells (YM4127) grown on media containing different carbon sources (2% galactose, 2% raffinose, and 4% glucose). (A and B) ChIP and immunoblot analyses of Rgt1 using anti-Rgt1 antibody were performed as described in the legends to Fig. 5 and 7A, respectively. The HXT promoters in the immunoprecipitated DNA (IP) were PCR amplified (A), quantified by the ImageQuaNT program, and presented as the ratio of immunoprecipitated counts to input counts (B). (C) Time course of dissociation of Rgt1 from the HXT promoters. Yeast cells (YM4127) grown on galactose (2%) (Gal) were harvested after glucose (4%) was added at the time points indicated above each lane and subjected to ChIP. After the HXT promoters in the immunoprecipitated DNA were amplified by PCR, the products were analyzed as described in the legend to Fig. 6. Chromatin prepared from the Δ_rgt1_ (YM4509) strain was used as the control for ChIP using anti-Rgt1 antibody (box). WT, wild type.
FIG. 9.
Glucose-induced phosphorylation inhibits DNA-binding activity of Rgt1 in vitro. (A) IDBA. LexA-Rgt1 was immunoprecipitated from yeast extracts as described in the legend to Fig. 7A and incubated with 32P-labeled DNA containing four Rgt1 binding sites (a repeat of Rgt1 binding sites C1 and C2). Approximately 85% of the DNA was bound to anti-LexA-Rgt1 beads. The bound DNA was eluted with 1 M NaCl. The Rgt1 bound to anti-LexA beads was subsequently eluted by boiling the beads in SDS buffer. The eluted DNA and Rgt1 were resolved in polyacrylamide gels and visualized by autoradiography and Western blotting (using anti-LexA antibody), respectively. (B) Rgt1 is phosphorylated. Rgt1 was immunoprecipitated as described above and incubated with (+) or without (−) 10 U of CIP at 37°C for 30 min and then subjected to Western blotting. (C) Glucose-induced phosphorylation of Rgt1 inhibits its DNA-binding activity. IDBA (DNA) and Western blotting (Rgt1) were performed on phosphorylated and dephosphorylated (CIP-treated) Rgt1 as for panels A and B.
FIG. 10.
Constitutive DNA binding by Rgt1 leads to constitutive repression. (A) Chromatin prepared from cells expressing wild type (WT) Rgt1 (pBM3580) and an Rgt1 derivative missing 10 amino acids (Δ750-760; pBM4058) was immunoprecipitated using anti-Rgt1 antibody, and the HXT3 promoter in the immunoprecipitated DNA (IP) was detected by PCR as described in the legend to Fig. 8C. (B) DNA binding of Rgt1 in vitro. IDBA (DNA) and Western blotting (Rgt1) were performed on the two forms of Rgt1 as described in the legend to Fig. 9.
FIG. 11.
Model for glucose induction of HXT gene expression. Hypophosphorylated Rgt1 is efficiently recruited and forms a stable repression complex with the Ssn6-Tup1 corepressor complex at multiple Rgt1 binding sites in the HXT promoters, leading to the repression of HXT gene expression. Rgt1 is hyperphosphorylated in response to glucose, causing its rapid dissociation from the HXT promoters, leading to induction of gene expression.
Similar articles
- How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose.
Polish JA, Kim JH, Johnston M. Polish JA, et al. Genetics. 2005 Feb;169(2):583-94. doi: 10.1534/genetics.104.034512. Epub 2004 Oct 16. Genetics. 2005. PMID: 15489524 Free PMC article. - DNA-binding properties of the yeast Rgt1 repressor.
Kim JH. Kim JH. Biochimie. 2009 Feb;91(2):300-3. doi: 10.1016/j.biochi.2008.09.002. Epub 2008 Oct 7. Biochimie. 2009. PMID: 18950675 Free PMC article. - Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae.
Kim JH, Johnston M. Kim JH, et al. J Biol Chem. 2006 Sep 8;281(36):26144-9. doi: 10.1074/jbc.M603636200. Epub 2006 Jul 14. J Biol Chem. 2006. PMID: 16844691 - Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1.
Lakshmanan J, Mosley AL, Ozcan S. Lakshmanan J, et al. Curr Genet. 2003 Oct;44(1):19-25. doi: 10.1007/s00294-003-0423-2. Epub 2003 Jul 9. Curr Genet. 2003. PMID: 14508605 - Function and regulation of yeast hexose transporters.
Ozcan S, Johnston M. Ozcan S, et al. Microbiol Mol Biol Rev. 1999 Sep;63(3):554-69. doi: 10.1128/MMBR.63.3.554-569.1999. Microbiol Mol Biol Rev. 1999. PMID: 10477308 Free PMC article. Review.
Cited by
- Asymmetric signal transduction through paralogs that comprise a genetic switch for sugar sensing in Saccharomyces cerevisiae.
Sabina J, Johnston M. Sabina J, et al. J Biol Chem. 2009 Oct 23;284(43):29635-43. doi: 10.1074/jbc.M109.032102. Epub 2009 Aug 31. J Biol Chem. 2009. PMID: 19720826 Free PMC article. - A fungal family of transcriptional regulators: the zinc cluster proteins.
MacPherson S, Larochelle M, Turcotte B. MacPherson S, et al. Microbiol Mol Biol Rev. 2006 Sep;70(3):583-604. doi: 10.1128/MMBR.00015-06. Microbiol Mol Biol Rev. 2006. PMID: 16959962 Free PMC article. Review. - Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae.
Kim JH, Brachet V, Moriya H, Johnston M. Kim JH, et al. Eukaryot Cell. 2006 Jan;5(1):167-73. doi: 10.1128/EC.5.1.167-173.2006. Eukaryot Cell. 2006. PMID: 16400179 Free PMC article. - Characterization of KlGRR1 and SMS1 genes, two new elements of the glucose signaling pathway of Kluyveromyces lactis.
Hnatova M, Wésolowski-Louvel M, Dieppois G, Deffaud J, Lemaire M. Hnatova M, et al. Eukaryot Cell. 2008 Aug;7(8):1299-308. doi: 10.1128/EC.00454-07. Epub 2008 Jun 13. Eukaryot Cell. 2008. PMID: 18552281 Free PMC article. - Nutrient Sensing at the Plasma Membrane of Fungal Cells.
Van Dijck P, Brown NA, Goldman GH, Rutherford J, Xue C, Van Zeebroeck G. Van Dijck P, et al. Microbiol Spectr. 2017 Mar;5(2):10.1128/microbiolspec.funk-0031-2016. doi: 10.1128/microbiolspec.FUNK-0031-2016. Microbiol Spectr. 2017. PMID: 28256189 Free PMC article. Review.
References
- Axelrod, J. D., M. S. Reagan, and J. Majors. 1993. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev. 7:857-869. - PubMed
- Baumgartner, U., B. Hamilton, M. Piskacek, H. Ruis, and H. Rottensteiner. 1999. Functional analysis of the Zn2Cys6 transcription factors Oaf1p and Pip2p: different roles in fatty acid induction of β-oxidation in Saccharomyces cerevisiae. J. Biol. Chem. 274:22208-22216. - PubMed
- Carlson, N. G., and J. W. Little. 1993. Highly cooperative DNA binding by Coliphage HK022 repressor. J. Mol. Biol. 230:1108-1130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases