NhaR and RcsB independently regulate the osmCp1 promoter of Escherichia coli at overlapping regulatory sites - PubMed (original) (raw)
NhaR and RcsB independently regulate the osmCp1 promoter of Escherichia coli at overlapping regulatory sites
Rachel Sturny et al. J Bacteriol. 2003 Aug.
Abstract
Transcription of the Escherichia coli osmC gene is induced by several stress conditions. osmC is expressed from two overlapping promoters, osmCp1 and osmCp2. The proximal promoter, osmCp2, is transcribed at the entry into the stationary phase by the sigma(s) sigma factor. The distal promoter, osmCp1, is activated by NhaR and RcsB. NhaR is a positive regulator of the LysR family and is known to be an activator of the nhaA gene encoding an Na(+)/H(+) antiporter. RcsB is the response regulator of the RcsCDB His-Asp phosphorelay signal transduction system. Genetic data indicated that activation of osmCp1 by both NhaR and RcsB requires the same short sequences upstream of the -35 region of the promoter. Accordingly, DNase I footprint analysis indicated that both activators protect an overlapping region close to the -35 box of the promoter and suggested that the regulatory effect is direct. Despite the overlap of the binding sites, each activator acts independent of the other and is specific for a particular stress. NhaR can stimulate osmCp1 in response to an osmotic signal even in the absence of RcsB. RcsB is responsible for the induction of osmCp1 by alteration of the cell envelope, even in the absence of NhaR. osmCp1 as an example of multiple-stress-responsive promoter is discussed in light of a comparison of the NhaR and RcsB target regions in the Enterobacteriaceae.
Figures
FIG. 1.
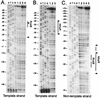
DNase I protection footprint of NhaR and RcsB in the osmCp1 region. A 188-bp DNA fragment encompassing the osmC promoter region, end labeled with 32P on the strand indicated at the bottom, was incubated with purified RcsB or RcsBD56E protein or with crude extracts enriched or not enriched in RcsB or NhaR protein, digested with DNase I, and analyzed on denaturing polyacrylamide gels. The results of autoradiography of the dried gels are shown. (A) Lane 1, DNA probe alone; lane 2, DNA probe incubated with purified RcsB protein (10 μM); lanes 3 and 4, DNA probe incubated with 100 μM (lane 3) or 10 μM (lane 4) purified RcsBD56E protein. (B) Lane 1, DNA probe alone; lanes 2 and 3, DNA probe incubated with crude extract (2 μg of protein) of strain CLG772 transformed with pAPT_nhaR_ (lane 2) or pAPT110 (lane 3). (C) Lane 1, DNA probe alone; lane 2, DNA probe incubated with crude extract (2 μg of protein) of strain CLG772 transformed with pAPT110; lane 3, DNA probe incubated with crude extract (2 μg of protein) of strain CLG772 transformed with pHRcsB; lane 4, DNA probe incubated with 100 μM purified RcsBD56E protein; lane 5, DNA probe incubated with crude extract (2 μg of protein) of strain CLG772 transformed with pAPT_nhaR_. The vertical lines and arrowheads indicate the positions protected from and hypersensitive to cleavage by DNase I, respectively.
FIG. 2.
Sequences of the RcsB and NhaR binding sites upstream of osmCp1. The sequence of the osmC promoter region is shown. Solid boxes indicate the −35 and −10 hexanucleotides of osmCp1. The dashed box indicates the −10 region of the proximal promoter osmCp2. Arrows indicate the transcription start sites. The osmCp11 and osmCp21 mutations, eliminating activity of osmCp1 and osmCp2, respectively, are indicated. The dots and arrowheads indicate the sites protected from and hypersensitive to cleavage by DNase I, respectively. The consensus sequence for RcsA-independent RcsB sites is shown above the RcsB sequence.
FIG. 3.
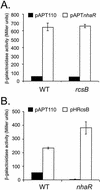
RcsB and NhaR activate an _osmCp1_-lacZ transcriptional fusion independent of each other. Strain CLG772 (WT), carrying an osmCp1_-lac fusion, and derivatives of this strain with mutations in rcsB (CLG805) (A) and nhaR (CLG806) (B) were transformed with plasmid pAPT110 (vector) (A and B), pAPT_nhaR (A), or pHRcsB (B). Overnight cultures of these strains in LB170 containing spectinomycin were diluted 1,000-fold and grown for five generations. They were then diluted 40-fold in prewarmed medium with IPTG (final concentration, 500 μM), and samples were used for _β_-galactosidase assays after 2 h. The values are the means of the results of three independent experiments.
FIG. 4.
Alignment of the DNA sequences of the osmCp regions of different members of the Enterobacteriaceae. MG1655, E. coli K-12 strain; CFT073, uropathogenic E. coli O6:H1 strain; EDL933, enterohemorrhagic E. coli O157:H7 strain; Shi fl, S. flexneri serotype 2a; Sal thy, S.enterica serovar Typhi CT18; Sal tm, S.enterica serovar Typhimurium LT2. The stars indicate differences compared with the MG1655 sequence. SD, Shine-Dalgarno sequence.
FIG. 5.
Alignment of putative RcsB target sites of different members of the Enterobacteriaceae: regulatory regions of the wza (A), _ftsA_1p (B), and rprA (C) genes. MG1655, E. coli K-12 strain; EDL933, enterohemorrhagic E. coli O157:H7 strain; Shi fl, S. flexneri serotype 2a; Sal thy, S.enterica serovar Typhi CT18; Sal tm, S.enterica serovar Typhimurium LT2. The stars indicate differences compared with the MG1655 sequence. The −10 and −35 regions of the promoters (shaded background) and the transcription start sites (boldface type) have been described previously (5, 18, 32).
Similar articles
- Multistress regulation in Escherichia coli: expression of osmB involves two independent promoters responding either to sigmaS or to the RcsCDB His-Asp phosphorelay.
Boulanger A, Francez-Charlot A, Conter A, Castanié-Cornet MP, Cam K, Gutierrez C. Boulanger A, et al. J Bacteriol. 2005 May;187(9):3282-6. doi: 10.1128/JB.187.9.3282-3286.2005. J Bacteriol. 2005. PMID: 15838058 Free PMC article. - The transcriptional activator NhaR is responsible for the osmotic induction of osmC(p1), a promoter of the stress-inducible gene osmC in Escherichia coli.
Toesca I, Perard C, Bouvier J, Gutierrez C, Conter A. Toesca I, et al. Microbiology (Reading). 2001 Oct;147(Pt 10):2795-2803. doi: 10.1099/00221287-147-10-2795. Microbiology (Reading). 2001. PMID: 11577158 - Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC.
Bouvier J, Gordia S, Kampmann G, Lange R, Hengge-Aronis R, Gutierrez C. Bouvier J, et al. Mol Microbiol. 1998 Jun;28(5):971-80. doi: 10.1046/j.1365-2958.1998.00855.x. Mol Microbiol. 1998. PMID: 9663683 - The molecular mechanism of regulation of the NhaA Na+/H+ antiporter of Escherichia coli, a key transporter in the adaptation to Na+ and H+.
Padan E, Gerchman Y, Rimon A, Rothman A, Dover N, Carmel-Harel O. Padan E, et al. Novartis Found Symp. 1999;221:183-96; discussion 196-9. doi: 10.1002/9780470515631.ch12. Novartis Found Symp. 1999. PMID: 10207920 Review. - The LeuO regulator and quiescence: About transcriptional roadblocks, multiple promoters, and crispr-cas.
Sánchez-Popoca D, Serrano-Fujarte I, Fernández-Mora M, Calva E. Sánchez-Popoca D, et al. Mol Microbiol. 2022 Nov;118(5):503-509. doi: 10.1111/mmi.14990. Epub 2022 Oct 20. Mol Microbiol. 2022. PMID: 36203248 Review.
Cited by
- Multistress regulation in Escherichia coli: expression of osmB involves two independent promoters responding either to sigmaS or to the RcsCDB His-Asp phosphorelay.
Boulanger A, Francez-Charlot A, Conter A, Castanié-Cornet MP, Cam K, Gutierrez C. Boulanger A, et al. J Bacteriol. 2005 May;187(9):3282-6. doi: 10.1128/JB.187.9.3282-3286.2005. J Bacteriol. 2005. PMID: 15838058 Free PMC article. - BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS.
Venkatesh GR, Kembou Koungni FC, Paukner A, Stratmann T, Blissenbach B, Schnetz K. Venkatesh GR, et al. J Bacteriol. 2010 Dec;192(24):6456-64. doi: 10.1128/JB.00807-10. Epub 2010 Oct 15. J Bacteriol. 2010. PMID: 20952573 Free PMC article. - The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine.
Goller C, Wang X, Itoh Y, Romeo T. Goller C, et al. J Bacteriol. 2006 Dec;188(23):8022-32. doi: 10.1128/JB.01106-06. Epub 2006 Sep 22. J Bacteriol. 2006. PMID: 16997959 Free PMC article. - A complex transcription network controls the early stages of biofilm development by Escherichia coli.
Prüss BM, Besemann C, Denton A, Wolfe AJ. Prüss BM, et al. J Bacteriol. 2006 Jun;188(11):3731-9. doi: 10.1128/JB.01780-05. J Bacteriol. 2006. PMID: 16707665 Free PMC article. Review. No abstract available. - The LysR-type transcriptional regulator VirR is required for expression of the virulence gene vapA of Rhodococcus equi ATCC 33701.
Russell DA, Byrne GA, O'Connell EP, Boland CA, Meijer WG. Russell DA, et al. J Bacteriol. 2004 Sep;186(17):5576-84. doi: 10.1128/JB.186.17.5576-5584.2004. J Bacteriol. 2004. PMID: 15317761 Free PMC article.
References
- Atichartpongkul, S., S. Loprasert, P. Vattanaviboon, W. Whangsuk, J. D. Helmann, and S. Mongkolsuk. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147:1775-1782. - PubMed
- Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- Bouvier, J., S. Gordia, G. Kampmann, R. Lange, R. Hengge-Aronis, and C. Gutierrez. 1998. Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol. Microbiol. 28:971-980. - PubMed
- Carballes, F., C. Bertrand, J. P. Bouche, and K. Cam. 1999. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol. Microbiol. 34:442-450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases