Phylogeny of east Asian mitochondrial DNA lineages inferred from complete sequences - PubMed (original) (raw)
Phylogeny of east Asian mitochondrial DNA lineages inferred from complete sequences
Qing-Peng Kong et al. Am J Hum Genet. 2003 Sep.
Erratum in
- Am J Hum Genet. 2004 Jun;75(1):157
Abstract
The now-emerging mitochondrial DNA (mtDNA) population genomics provides information for reconstructing a well-resolved mtDNA phylogeny and for discerning the phylogenetic status of the subcontinentally specific haplogroups. Although several major East Asian mtDNA haplogroups have been identified in studies elsewhere, some of the most basal haplogroups, as well as numerous minor subhaplogroups, were not yet determined or fully characterized. To fill the lacunae, we selected 48 mtDNAs from >2,000 samples across China for complete sequencing that cover virtually all (sub)haplogroups discernible to date in East Asia. This East Asian mtDNA phylogeny can henceforth serve as a solid basis for phylogeographic analyses of mtDNAs, as well as for studies of mitochondrial diseases in East and Southeast Asia.
Figures
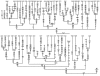
Figure 1
Phylogenetic tree of 48 East Asian mtDNA lineages, which were sampled from various regional Han populations (Yao et al. 2002_a_, 2003_a_), except for those with prefixes DW (Daur, from Inner Mongolia), EWK (Ewenki, from Inner Mongolia), Mg (Mongolian, from Inner Mongolia), and Miao (Miaozu, from Hunan). This tree incorporates the information drawn from previous reports (Derbeneva et al. ; Herrnstadt et al. ; Kivisild et al. , and references therein; Mishmar et al. 2003) by indicating the roots of several haplogroups from which those Asian sequences branch off. Also indicated are the Native American haplogroups A2, B2, C1, D1 (Bandelt et al., in press) and D2, D3 (Derbeneva et al. 2002), of which only C1 and D2 are found in Asia, too. The mutations (reconstructed most parsimoniously) on uninterrupted branches are listed (arbitrarily) in ascending order. Suffixes A, C, G, and T indicate transversion, “d” indicates a deletion, and a plus sign (+) indicates an insertion; the recurrent mutations are underlined. The reconstruction of the mutations at site 16519 and deletions of CA repeats around sites 522–523, which are known to be extremely variable, is tentative at best. Note that 2232+A, one of the specific mutations of haplogroup C4, was inadvertently scored as 2233+A in figure 1 of Kivisild et al. (2002). Further, the insertion defining haplogroup M7a1 is now scored as 5899+C.
Similar articles
- The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes.
Sun C, Kong QP, Palanichamy MG, Agrawal S, Bandelt HJ, Yao YG, Khan F, Zhu CL, Chaudhuri TK, Zhang YP. Sun C, et al. Mol Biol Evol. 2006 Mar;23(3):683-90. doi: 10.1093/molbev/msj078. Epub 2005 Dec 16. Mol Biol Evol. 2006. PMID: 16361303 - Genetic structure of Hmong-Mien speaking populations in East Asia as revealed by mtDNA lineages.
Wen B, Li H, Gao S, Mao X, Gao Y, Li F, Zhang F, He Y, Dong Y, Zhang Y, Huang W, Jin J, Xiao C, Lu D, Chakraborty R, Su B, Deka R, Jin L. Wen B, et al. Mol Biol Evol. 2005 Mar;22(3):725-34. doi: 10.1093/molbev/msi055. Epub 2004 Nov 17. Mol Biol Evol. 2005. PMID: 15548747 - Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations.
Kong QP, Bandelt HJ, Sun C, Yao YG, Salas A, Achilli A, Wang CY, Zhong L, Zhu CL, Wu SF, Torroni A, Zhang YP. Kong QP, et al. Hum Mol Genet. 2006 Jul 1;15(13):2076-86. doi: 10.1093/hmg/ddl130. Epub 2006 May 19. Hum Mol Genet. 2006. PMID: 16714301 - Ancient mitochondrial DNA from Malaysian hair samples: some indications of Southeast Asian population movements.
Ricaut FX, Bellatti M, Lahr MM. Ricaut FX, et al. Am J Hum Biol. 2006 Sep-Oct;18(5):654-67. doi: 10.1002/ajhb.20535. Am J Hum Biol. 2006. PMID: 16917897 - Contribution of Mitochondrial DNA Variation to Chronic Disease in East Asian Populations.
Sun D, Wei Y, Zheng HX, Jin L, Wang J. Sun D, et al. Front Mol Biosci. 2019 Nov 15;6:128. doi: 10.3389/fmolb.2019.00128. eCollection 2019. Front Mol Biosci. 2019. PMID: 31803756 Free PMC article. Review.
Cited by
- Effective strategies for forensic analysis in the mitochondrial DNA coding region.
Coble MD, Vallone PM, Just RS, Diegoli TM, Smith BC, Parsons TJ. Coble MD, et al. Int J Legal Med. 2006 Jan;120(1):27-32. doi: 10.1007/s00414-005-0044-z. Epub 2005 Oct 28. Int J Legal Med. 2006. PMID: 16261373 - Beringian standstill and spread of Native American founders.
Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, Mulligan CJ, Bravi CM, Rickards O, Martinez-Labarga C, Khusnutdinova EK, Fedorova SA, Golubenko MV, Stepanov VA, Gubina MA, Zhadanov SI, Ossipova LP, Damba L, Voevoda MI, Dipierri JE, Villems R, Malhi RS. Tamm E, et al. PLoS One. 2007 Sep 5;2(9):e829. doi: 10.1371/journal.pone.0000829. PLoS One. 2007. PMID: 17786201 Free PMC article. - The impact of mitochondrial DNA and nuclear genes related to mitochondrial functioning on the risk of Parkinson's disease.
Gaweda-Walerych K, Zekanowski C. Gaweda-Walerych K, et al. Curr Genomics. 2013 Dec;14(8):543-59. doi: 10.2174/1389202914666131210211033. Curr Genomics. 2013. PMID: 24532986 Free PMC article. - Genetic imprint of the Mongol: signal from phylogeographic analysis of mitochondrial DNA.
Cheng B, Tang W, He L, Dong Y, Lu J, Lei Y, Yu H, Zhang J, Xiao C. Cheng B, et al. J Hum Genet. 2008;53(10):905-913. doi: 10.1007/s10038-008-0325-8. Epub 2008 Sep 4. J Hum Genet. 2008. PMID: 18769869 - Is mitochondrial tRNA(phe) variant m.593T>C a synergistically pathogenic mutation in Chinese LHON families with m.11778G>A?
Zhang AM, Bandelt HJ, Jia X, Zhang W, Li S, Yu D, Wang D, Zhuang XY, Zhang Q, Yao YG. Zhang AM, et al. PLoS One. 2011;6(10):e26511. doi: 10.1371/journal.pone.0026511. Epub 2011 Oct 19. PLoS One. 2011. PMID: 22039503 Free PMC article.
References
Electronic-Database Information
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the mtDNA complete sequence data [accession numbers AY255133–AY255180])
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 - PubMed
- Bandelt H-J, Herrnstadt C, Yao Y-G, Kong Q-P, Kivisild T, Rengo C, Scozzari R, Richards M, Villems R, Macaulay V, Howell N, Torroni A, Zhang Y-P. Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet (in press) - PubMed
- Bandelt H-J, Lahermo P, Richards M, Macaulay V (2001) Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med 115:64–69 - PubMed
- De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafé M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C (1999) Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J 13:1532–1536 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases