Interaction between Ski7p and Upf1p is required for nonsense-mediated 3'-to-5' mRNA decay in yeast - PubMed (original) (raw)
Interaction between Ski7p and Upf1p is required for nonsense-mediated 3'-to-5' mRNA decay in yeast
Shinya Takahashi et al. EMBO J. 2003.
Abstract
Aberrant mRNAs containing premature termination codons (PTC-mRNAs) are degraded by a conserved surveillance system, referred to as the nonsense- mediated decay (NMD) pathway. Although NMD is reported to operate on the decapping and 5'-to-3' exonucleolytic decay of PTC-mRNAs without affecting deadenylation, a role for an opposite 3'-to-5' decay pathway remains largely unexplored. In this study, we have characterized the 3'-to-5' directed mRNA degradation in the yeast NMD pathway. PTC-mRNAs are stabilized in yeast cells lacking the components of 3'-to-5' mRNA-decay machinery. The 3'-to-5' directed degradation of PTC-mRNAs proceeds more rapidly than that of the PTC-free transcript, in a manner dependent on the cytoplasmic exosome and Upf proteins. Moreover, Upf1p, but not Upf2p, interacts physically with an N-terminal domain of Ski7p, although the interaction requires Upf2p. The efficiency of 3'-to-5' directed degradation of PTC-mRNAs is impaired by overexpression of Ski7p N-domain fragments that contain a sequence of the Upf1p-interaction region. These data suggest that the activation of 3'-to-5' directed NMD is mediated through the interaction between Upf1p and the Ski7p N domain.
Figures
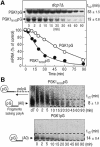
Fig. 1. Yeast strains lacking the components of 3′-to-5′ mRNA-decay machinery accumulate an aberrant mRNA containing PTC, PGK1N103pG. (A) Yeast strains, wild type (WT, YS001), _ski7_Δ (YS002), _ski7/C_Δ (YS063), _ski2_Δ (YS060), _ski3_Δ (YS061), _ski8_Δ (YS014), rrp4-1 (YS094) and _upf1_Δ (YS065), which carry the PGK1N103pG reporter (pRP611), were grown at 26°C in a galactose-containing medium and harvested at mid-log phase. The aberrant mRNA was detected by northern blot analysis with end-labeled oligonucleotides specific for the PGK1N103pG (upper panel). The signals were quantified using a PhosphorImager and corrected for loading using the SRP RNA, scR1 (lower panel). The values under PGK1N103pG northern blot show relative intensity to the amount of mRNA obtained in the WT strain. (B) Yeast strains, wild type (WT, YS001), _ski7_Δ (YS002) and _xrn1_Δ (YS065), which carry the PGK1N103pG reporter (pRP611), were grown to mid-log phase in the galactose medium and shifted to a glucose medium to repress transcription of the reporter mRNA. Time points represent minutes after transcriptional repression. The half-lives (t1/2; min) of PGK1N103pG mRNA are shown as means ± standard deviations (SD), which were obtained from at least three independent experiments.
Fig. 2. 3′-to-5′ mRNA degradation is accelerated in the aberrant mRNA containing PTC. (A) Yeast _dcp1_Δ strain (YS055) carrying PGK1pG (top panel and open circles) or PGK1N103pG (bottom panel and closed circles) reporters was grown to mid-log phase in the galactose medium and shifted to a glucose medium to repress transcription of the reporter mRNAs. After incubation for the indicated times, northern blot analysis was performed as described in Figure 1, and the amounts of transcript are illustrated as the function of the times. (B) Wild-type strain (YS001) that carries PGK1 N103pG (top panel) or PGK1pG (bottom panel) reporter was grown in a minimal medium containing galactose and shifted to the medium supplemented with glucose and cycloheximide to inhibit the transcription and decapping. After incubation for the indicated times, northern blot analysis was performed as described in Figure 1. The sample from the 0-min time that had been cleaved by RNaseH with oligo dT was also applied to the upper left lane (dT), which indicates the position of poly(G)–3′-end fragments lacking poly(A). The half-lives (t1/2; min) of mRNAs [full-length forms and fragments lacking poly(A) tail for A and B, respectively] are shown as means ± SD, which were obtained from three independent experiments.
Fig. 3. 3′-to-5′ directed NMD is mediated through the recognition of PTC and requires both the exosome and the Ski proteins. (A) Yeast _xrn1_Δ gst1-1 (YS106) strains that carry PGK1N103pG reporter and either YCplac22-GST1 (bottom panel) or the empty vector (top panel) were grown at 25°C in the galactose medium to mid-log phase and incubated at 37°C for 1.5 h. (B) Yeast strains lacking the DCP1 gene (_dcp1_Δ, YS055; _dcp1_Δ rrp4-1, YS095) or containing its temperature-sensitive allele (dcp1-2, YS088; _dcp11-2 ski2_Δ, YS089; _dcp1-2 ski3_Δ, YS090; _dcp1-2 ski7_Δ, YS091; _dcp1-2 ski8_Δ, YS092) which carry the PGK1N103pG reporter were grown at 25°C in the galactose medium to mid-log phase and incubated at 37°C for 1.5 h. Transcription was repressed by the addition of glucose. After incubation for the indicated times, northern blot analysis was performed as described in Figure 1. The half-lives (t1/2; min) of mRNAs are shown as means ± SD, which were obtained from at least three independent experiments.
Fig. 4. Acceleration of 3′-to-5′ directed NMD is mediated through Upf proteins. Yeast strains lacking XRN1 and/or UPF genes (_xrn1_Δ, YS064; _xrn1_Δ _upf1_Δ, YS070; _xrn1_Δ _upf2_Δ YS071; _xrn1_Δ _upf3_Δ, YS072), which carry PGK1N103pG (A) and PGK1pG (B) reporters, were grown at 25°C in the galactose medium to mid-log phase. Transcriptional repression and hybridization were performed as described for Figure 1. The half-lives (t1/2; min) of mRNAs are shown as means ± SD, which were obtained from at least three independent experiments.
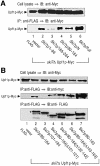
Fig. 5. The N-terminal domain of Ski7p associates with Upf1p in a manner dependent on Upf2p. Cell extracts were prepared from _SKI7_- and/or _UPF1/2_-deletion strains that carry Upf1p-Myc (A; lanes 1–4, 9–12 for YS037–YS040 and lanes 5–8 for YS045–YS048) or Upf2p-Myc (B; lanes 1–4, 9–12 for YS041–YS044 and lanes 5–8 for YS049–YS052). The yeast strains also expressed the indicated forms of FLAG-tagged Ski7p. The extracts (lanes 1–8) were directly subjected to immunoprecipitation (IP) with an anti-FLAG. In some experiments, the extracts that had been treated with RNase were also subjected to the immunoprecipitation assay (lanes 9–12, RNase-treated). The precipitated proteins (bottom panels), together with the whole-cell extracts (top panels), were immunoblotted (IB) with an anti-Myc (9E10) antibody to detect Upf1p-Myc and Upf2p-Myc.
Fig. 6. Mapping of Upf1p-binding sites on the N-terminal domain of Ski7p. Cell extracts were prepared from yeast strains expressing Upf1p-Myc and the indicated forms of FLAG-tagged Ski7p (A, lanes 1–6 for YS037, YS101, YS103, YS104, YS038, YS039; and B, lanes 1–7 for YS037, YS038, YS123, YS124, YS125, YS126, YS127). The extracts were immunoprecipitated (IP) with the anti-FLAG antibody. The precipitated proteins (bottom panel) and whole-cell extracts (top panel) were immunoblotted (IB) with the anti-Myc antibody to detect Upf1p-Myc. The asterisks in (B) indicate FLAG-tagged Ski7p mutants expressed in the various strains.
Fig. 7. Interaction of the N-terminal domain of Ski7p with Upf1p is required for 3′-to-5′ directed NMD. The _xrn1_Δ strains expressing PGK1N103pG (A) and PGK1pG (B) reporters were transformed with plasmids carrying the indicated deletion mutants of Ski7p/N domain (vector, YS077; Ski7p/1–96, YS082; Ski7p/80–264, YS083; Ski7p/80–184, YS085; Ski7p/168–264, YS084; Ski7p/101–184, YS128; Ski7p/80–163; YS132). The transformants were grown in the galactose- containing medium and harvested at mid-log phase. Transcriptional repression and hybridization were performed as described for Figure 1. The half-lives (t1/2; min) of mRNAs are shown as means ± SD, which were obtained from at least three independent experiments.
Fig. 8. The N domain of Ski7p interacting with Upf1p. Deletion mutants of Ski7p used in this study are shown, together with the summary of present results obtained in Figures 6 and 7.
Similar articles
- An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3'-->5' degradation.
Mitchell P, Tollervey D. Mitchell P, et al. Mol Cell. 2003 May;11(5):1405-13. doi: 10.1016/s1097-2765(03)00190-4. Mol Cell. 2003. PMID: 12769863 - Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover.
Weng Y, Czaplinski K, Peltz SW. Weng Y, et al. Mol Cell Biol. 1996 Oct;16(10):5491-506. doi: 10.1128/MCB.16.10.5491. Mol Cell Biol. 1996. PMID: 8816462 Free PMC article. - Targeting of aberrant mRNAs to cytoplasmic processing bodies.
Sheth U, Parker R. Sheth U, et al. Cell. 2006 Jun 16;125(6):1095-109. doi: 10.1016/j.cell.2006.04.037. Cell. 2006. PMID: 16777600 Free PMC article. - NMD monitors translational fidelity 24/7.
Celik A, He F, Jacobson A. Celik A, et al. Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review. - Cutting the nonsense: the degradation of PTC-containing mRNAs.
Nicholson P, Mühlemann O. Nicholson P, et al. Biochem Soc Trans. 2010 Dec;38(6):1615-20. doi: 10.1042/BST0381615. Biochem Soc Trans. 2010. PMID: 21118136 Review.
Cited by
- The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome.
Halbach F, Rode M, Conti E. Halbach F, et al. RNA. 2012 Jan;18(1):124-34. doi: 10.1261/rna.029553.111. Epub 2011 Nov 23. RNA. 2012. PMID: 22114319 Free PMC article. - A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay.
Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. Cole SE, et al. Mol Cell. 2009 May 14;34(4):440-50. doi: 10.1016/j.molcel.2009.04.017. Mol Cell. 2009. PMID: 19481524 Free PMC article. - Cytoplasmic foci are sites of mRNA decay in human cells.
Cougot N, Babajko S, Séraphin B. Cougot N, et al. J Cell Biol. 2004 Apr;165(1):31-40. doi: 10.1083/jcb.200309008. Epub 2004 Apr 5. J Cell Biol. 2004. PMID: 15067023 Free PMC article. - Coupled protein quality control during nonsense-mediated mRNA decay.
Inglis AJ, Guna A, Gálvez-Merchán Á, Pal A, Esantsi TK, Keys HR, Frenkel EM, Oania R, Weissman JS, Voorhees RM. Inglis AJ, et al. J Cell Sci. 2023 May 15;136(10):jcs261216. doi: 10.1242/jcs.261216. Epub 2023 May 23. J Cell Sci. 2023. PMID: 37218462 Free PMC article. - Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome.
Orban TI, Izaurralde E. Orban TI, et al. RNA. 2005 Apr;11(4):459-69. doi: 10.1261/rna.7231505. Epub 2005 Feb 9. RNA. 2005. PMID: 15703439 Free PMC article.
References
- Atkin A.L., Schenkman,L.R., Eastham,M., Dahlseid,J.N., Lelivelt,M.J. and Culbertson,M.R. (1997) Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem., 272, 22163–22172. - PubMed
- Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature, 382, 642–646. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases