Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways - PubMed (original) (raw)
Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways
David Gatfield et al. EMBO J. 2003.
Abstract
The nonsense-mediated mRNA decay (NMD) pathway promotes the rapid degradation of mRNAs containing premature stop codons (PTCs). In Caenorhabditis elegans, seven genes (smg1-7) playing an essential role in NMD have been identified. Only SMG2-4 (known as UPF1-3) have orthologs in Saccharomyces cerevisiae. Here we show that the Drosophila orthologs of UPF1-3, SMG1, SMG5 and SMG6 are required for the degradation of PTC-containing mRNAs, but that there is no SMG7 ortholog in this organism. In contrast, orthologs of SMG5-7 are encoded by the human genome and all three are required for NMD. In human cells, exon boundaries have been shown to play a critical role in defining PTCs. This role is mediated by components of the exon junction complex (EJC). Contrary to expectation, however, we show that the components of the EJC are dispensable for NMD in Drosophila cells. Consistently, PTC definition occurs independently of exon boundaries in Drosophila. Our findings reveal that despite conservation of the NMD machinery, different mechanisms have evolved to discriminate premature from natural stop codons in metazoa.
Figures
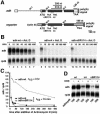
Fig. 1. Generation of an NMD reporter for Drosophila cells. (A) Schematic representation of the adh gene and the reporter used in this study. Black boxes, exons; white boxes, adh 5′- or 3′-UTRs; gray boxes, sequences derived from vector pAc5.1b; IN, introns. The PTC-containing constructs correspond to adh-nBR114 and adh-n4 (Brogna, 1999). (B) S2 cells constitutively expressing adh-wt or the PTC reporters were incubated with actinomycin D (5 µg/ml) for the times indicated above the lanes. Total RNA samples were isolated and analyzed by northern blot using probes specific for adh and rp49 mRNAs. (C) The levels of adh-wt, adh-n4 and adh-nBR114 were quantitated, normalized to the levels of rp49 mRNA (whose levels do not change relative to 18S rRNA within the time frame of the experiment) and plotted as a function of time. The half-lives of the mRNAs are indicated. (D) S2 cells constitutively expressing adh-wt or the PTC reporters were incubated with cycloheximide (CHX, 100 µg/ml) for 45 min. Total RNA samples were analyzed by northern blot. The numbers below the lanes indicate the levels of adh transcripts normalized to those of rp49 mRNA. These values were set to 100% in untreated cells.
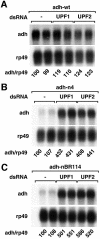
Fig. 2. Adh reporters are subjected to NMD in Drosophila cells. (A–C) S2 cell lines expressing adh-wt, adh-n4 or adh-nBR114 were transfected with dsRNAs corresponding to UPF1 or UPF2 as indicated. Transfections were carried out in duplicate. Total RNA samples were analyzed by northern blot as described in Figure 1.
Fig. 3. Phylogenetic tree and domain organization of the SMG5–7 proteins. (A) The two identified orthologous groups are shown in red. Only one and a half TPR repeats can be identified automatically and are indicated in dark blue within the conserved helical N-terminal domain (cyan). The C.elegans sequences are underlined and are known to be involved in NMD (Cali et al., 1999; Page et al., 1999; Anders et al., 2003). Human and Drosophila sequences characterized in this study are shown in gray boxes. The vertical black bars on the schematic of the protein domains indicate that part of the protein sequence is not drawn to scale. The number of amino acid residues is shown on the right. Scale bar: 200 amino acids. TPR, tetratricopeptide repeats; PIN, PilT-N-terminus; Ce, Caenorhabditis elegans; Ci, Ciona intestinalis; Dm, Drosophila melanogaster; Fr, Fugu rubripes; Hs, Homo sapiens. (B) Multiple sequence alignment of the TPR-like region. The accession number of each sequence is indicated, as well as the species and the start and end points of the sequences. For the sequences of C.intestinalis and F.rubripes, no accession numbers are shown, as the predictions were performed directly on the genomic sequences using GeneWise (Birney et al., 1996). The consensus in 70% of the sequences is shown below the multialignment; h, l, p, +, – represent hydrophobic, aliphatic, polar, positive and negative residues, respectively. Aliphatic residues are highlighted in cyan, hydrophobic in blue, polar in green, negative in pink, and positive in red. The conserved glycines are highlighted in orange. The secondary structure prediction (Sec.Str.Pred.) is taken from the consensus of the multiple alignment (H, helices predicted with an accuracy >82%; h, helices predicted with an accuracy ≤82%]. The blue bars above the sequences show the position of the one and a half TPR repeats identified by Clissold and Ponting (2000).
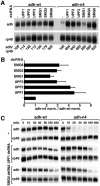
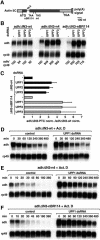
Fig. 4. SMG1, SMG5 and SMG6 are essential for NMD in Drosophila cells. (A) S2 cell lines expressing adh-wt or adh-n4 were transfected with the dsRNAs indicated above the lanes. RNA samples were analyzed by northern blot, as described in Figure 2. (B) The steady-state levels of adh-wt and adh-n4 mRNAs were quantitated in at least three independent experiments and normalized to those of rp49 mRNA. For each experiment, the normalized values (norm.) obtained for n4 mRNA were divided by those obtained for adh-wt, to correct for potential non-specific effects of the depletions. This ratio was set arbitrarily to a value of 1 in control cells. The mean values ± SDs are shown. (C) S2 cell lines expressing adh-wt or adh-n4 were transfected with dsRNAs specific for UPF1 and SMG6. Seven days after addition of dsRNAs, treated and untreated cells were incubated with actinomycin D (5 µg/ml) for the times indicated above the lanes. Total RNA samples were isolated and analyzed by northern blot as described in Figure 1D.
Fig. 5. Human SMG5–7 are required for NMD in HeLa cells. (A) HeLa cells were transfected with the indicated siRNAs and plasmids expressing either β-globin-wt or β-globin-NS39 mRNAs. A plasmid expressing GFP–NXF1 was co-transfected to correct for differences in transfection efficiencies. RNA samples were analyzed by northern blot using β-globin and GFP probes. The numbers below the lanes indicate the levels of β-globin-wt or NS39 transcripts normalized to those of GFP–NXF1 mRNA and set to 100% in cells treated with an unrelated siRNA (–). (B) The steady-state levels of β-globin-wt or NS39 mRNAs were quantitated in at least three independent experiments and normalized to those of the GFP–NXF1 mRNA. The normalized values (norm.) obtained for NS39 were divided by those obtained for the wild-type mRNA. The mean values ± SDs are shown.
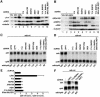
Fig. 6. The individual components of the EJC are dispensable for NMD in Drosophila. (A and B) S2 cells expressing adh-wt or adh-n4 were transfected with dsRNA specific for Y14, RNPS1, REF1, SRm160 or DEK, or with a mixture of dsRNAs (lanes 9 and 10) as indicated. Protein samples from total lysates of untreated or depleted cells (adh-n4 cell line) were analyzed by western blot using antibodies described in Gatfield and Izaurralde (2002). The anti-SRm160 antibody cross-reacts with three bands that are all depleted in cells treated with SRm160 dsRNA, suggesting that these bands may represent modified and/or alternatively spliced forms of the protein. Tubulin served as a loading control. In lanes 1–3, dilutions of the samples isolated on day 0 were loaded to estimate the efficiency of the depletion. (C and D) RNA samples isolated from depleted cells shown in (A) and (B) were isolated and analyzed by northern blot as described in Figure 2. Samples isolated from cells treated with UPF1 or UPF2 dsRNAs were analyzed in parallel. The numbers below the lanes indicate the levels of adh transcripts normalized to those of rp49 mRNA. (E) Normalized values obtained in three independent experiments for n4 were divided by those obtained for adh-wt. These ratios were set arbitrarily to 1 in control cells. (F) S2 cells expressing adh-n4 were transfected with dsRNA specific for UPF1, or with mixtures of dsRNAs as indicated. RNA samples were analyzed by northern blot. The numbers below the lanes indicate the levels of adh-n4 transcript normalized to those of rp49 mRNA.
Fig. 7. PTCs are defined independently of downstream exon boundaries in Drosophila cells. (A) Schematic representation of the reporters lacking a downstream intron. Symbols are as in Figure 1A. (B) S2 cells expressing adh-wt, adh-n4 or adh-nBR114 reporters lacking intron 3 (adhΔIN3-wt, adhΔIN3-n4, adhΔIN3-nBR114) were treated with UPF1, UPF2 or UPF3 dsRNAs. Total RNA samples were analyzed as described in Figure 2. (C) The steady-state levels of adhΔIN3-n4 and adhΔIN3-nBR114 mRNAs were quantitated in at least three independent experiments and normalized to those of rp49 mRNA. These normalized values (adhΔIN3-PTC norm.) were divided by those obtained for adhΔIN3-wt. The mean values ± SDs are shown. (D–F) S2 cells constitutively expressing adhΔIN3-wt, adhΔIN3-n4 or adhΔIN3-nBR114 were treated with UPF1 dsRNA. Seven days after addition of UPF1 dsRNA, treated or untreated cells (control) were incubated with actinomycin D (5 µg/ml) for the times indicated above the lanes. Total RNA samples were isolated and analyzed by northern blot.
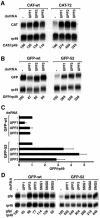
Fig. 8. PTCs can be recognized on a heterologous mRNA. (A) S2 cells expressing CAT-wt or CAT-72 mRNAs were treated with UPF1, UPF2 or UPF3 dsRNAs. Total RNA samples were analyzed by northern blot using probes specific for CAT and rp49 mRNAs. The numbers below the lanes indicate the levels of the CAT transcripts normalized to those of rp49 mRNA. (B) S2 cells expressing GFP-wt or GFP-52 mRNAs were treated with UPF1, UPF2 or UPF3 dsRNAs. The levels of the GFP transcripts normalized to those of rp49 mRNA are indicated below the lanes. (C) The levels of GFP-wt and GFP-52 mRNAs were quantitated in at least three independent experiments and normalized to those of rp49 mRNA. These ratios were set arbitrarily to 1 in control cells. The mean values ± SDs are shown. (D) S2 cell lines expressing GFP-wt or GFP-52 were treated with the dsRNAs indicated above the lanes. Total RNA samples were analyzed as described in (B).
Similar articles
- Multiple Nonsense-Mediated mRNA Processes Require Smg5 in Drosophila.
Nelson JO, Förster D, Frizzell KA, Luschnig S, Metzstein MM. Nelson JO, et al. Genetics. 2018 Aug;209(4):1073-1084. doi: 10.1534/genetics.118.301140. Epub 2018 Jun 14. Genetics. 2018. PMID: 29903866 Free PMC article. - Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways.
Metze S, Herzog VA, Ruepp MD, Mühlemann O. Metze S, et al. RNA. 2013 Oct;19(10):1432-48. doi: 10.1261/rna.038893.113. Epub 2013 Aug 20. RNA. 2013. PMID: 23962664 Free PMC article. - Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival.
Wittkopp N, Huntzinger E, Weiler C, Saulière J, Schmidt S, Sonawane M, Izaurralde E. Wittkopp N, et al. Mol Cell Biol. 2009 Jul;29(13):3517-28. doi: 10.1128/MCB.00177-09. Epub 2009 May 4. Mol Cell Biol. 2009. PMID: 19414594 Free PMC article. - Cutting the nonsense: the degradation of PTC-containing mRNAs.
Nicholson P, Mühlemann O. Nicholson P, et al. Biochem Soc Trans. 2010 Dec;38(6):1615-20. doi: 10.1042/BST0381615. Biochem Soc Trans. 2010. PMID: 21118136 Review. - Execution of nonsense-mediated mRNA decay: what defines a substrate?
Rebbapragada I, Lykke-Andersen J. Rebbapragada I, et al. Curr Opin Cell Biol. 2009 Jun;21(3):394-402. doi: 10.1016/j.ceb.2009.02.007. Epub 2009 Apr 7. Curr Opin Cell Biol. 2009. PMID: 19359157 Review.
Cited by
- UPF1-Mediated RNA Decay-Danse Macabre in a Cloud.
Lavysh D, Neu-Yilik G. Lavysh D, et al. Biomolecules. 2020 Jul 4;10(7):999. doi: 10.3390/biom10070999. Biomolecules. 2020. PMID: 32635561 Free PMC article. Review. - Zinc finger protein Zn72D promotes productive splicing of the maleless transcript.
Worringer KA, Panning B. Worringer KA, et al. Mol Cell Biol. 2007 Dec;27(24):8760-9. doi: 10.1128/MCB.01415-07. Epub 2007 Oct 8. Mol Cell Biol. 2007. PMID: 17923683 Free PMC article. - Selective profiling of ribosomes associated with yeast Upf proteins.
Ganesan R, Leszyk J, Jacobson A. Ganesan R, et al. Methods. 2019 Feb 15;155:58-67. doi: 10.1016/j.ymeth.2018.12.008. Epub 2018 Dec 26. Methods. 2019. PMID: 30593864 Free PMC article. - Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase.
Redon S, Reichenbach P, Lingner J. Redon S, et al. Nucleic Acids Res. 2007;35(20):7011-22. doi: 10.1093/nar/gkm724. Epub 2007 Oct 16. Nucleic Acids Res. 2007. PMID: 17940095 Free PMC article. - Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets.
Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Rehwinkel J, et al. RNA. 2005 Oct;11(10):1530-44. doi: 10.1261/rna.2160905. RNA. 2005. PMID: 16199763 Free PMC article.
References
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. - PubMed
- Benting J., Lecat,S., Zacchetti,D. and Simons,K. (2000) Protein expression in Drosophila Schneider cells. Anal. Biochem., 278, 59–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials