Statistical significance for genomewide studies - PubMed (original) (raw)
Statistical significance for genomewide studies
John D Storey et al. Proc Natl Acad Sci U S A. 2003.
Abstract
With the increase in genomewide experiments and the sequencing of multiple genomes, the analysis of large data sets has become commonplace in biology. It is often the case that thousands of features in a genomewide data set are tested against some null hypothesis, where a number of features are expected to be significant. Here we propose an approach to measuring statistical significance in these genomewide studies based on the concept of the false discovery rate. This approach offers a sensible balance between the number of true and false positives that is automatically calibrated and easily interpreted. In doing so, a measure of statistical significance called the q value is associated with each tested feature. The q value is similar to the well known p value, except it is a measure of significance in terms of the false discovery rate rather than the false positive rate. Our approach avoids a flood of false positive results, while offering a more liberal criterion than what has been used in genome scans for linkage.
Figures
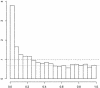
Fig. 1.
A density histogram of the 3,170 p values from the Hedenfalk_et al._ (14) data. The dashed line is the density histogram we would expect if all genes were null (not differentially expressed). The dotted line is at the height of our estimate of the proportion of null p values.
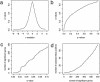
Fig. 2.
Results from the Hedenfalk et al. (14) data. (a) The_q_ values of the genes versus their respective t statistics. (b) The q values versus their respective p values. (c) The number of genes occurring on the list up through each_q_ value versus the respective q value. (d) The expected number of false positive genes versus the total number of significant genes given by the q values.
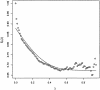
Fig. 3.
The versus λ for the data of Hedenfalk et al. (14). The solid line is a natural cubic spline fit to these points to estimate
.
Similar articles
- The false discovery rate: a key concept in large-scale genetic studies.
Chen JJ, Roberson PK, Schell MJ. Chen JJ, et al. Cancer Control. 2010 Jan;17(1):58-62. doi: 10.1177/107327481001700108. Cancer Control. 2010. PMID: 20010520 - Efficient computation of significance levels for multiple associations in large studies of correlated data, including genomewide association studies.
Dudbridge F, Koeleman BP. Dudbridge F, et al. Am J Hum Genet. 2004 Sep;75(3):424-35. doi: 10.1086/423738. Epub 2004 Jul 19. Am J Hum Genet. 2004. PMID: 15266393 Free PMC article. - Rank order metrics for quantifying the association of sequence features with gene regulation.
Clarke ND, Granek JA. Clarke ND, et al. Bioinformatics. 2003 Jan 22;19(2):212-8. doi: 10.1093/bioinformatics/19.2.212. Bioinformatics. 2003. PMID: 12538241 - Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments.
Johnson JM, Edwards S, Shoemaker D, Schadt EE. Johnson JM, et al. Trends Genet. 2005 Feb;21(2):93-102. doi: 10.1016/j.tig.2004.12.009. Trends Genet. 2005. PMID: 15661355 Review. - Bioinformatics analysis of alternative splicing.
Lee C, Wang Q. Lee C, et al. Brief Bioinform. 2005 Mar;6(1):23-33. doi: 10.1093/bib/6.1.23. Brief Bioinform. 2005. PMID: 15826354 Review.
Cited by
- A multi-regional human brain atlas of chromatin accessibility and gene expression facilitates promoter-isoform resolution genetic fine-mapping.
Dong P, Song L, Bendl J, Misir R, Shao Z, Edelstien J, Davis DA, Haroutunian V, Scott WK, Acker S, Lawless N, Hoffman GE, Fullard JF, Roussos P. Dong P, et al. Nat Commun. 2024 Nov 22;15(1):10113. doi: 10.1038/s41467-024-54448-y. Nat Commun. 2024. PMID: 39578476 Free PMC article. - Association of inflammatory cytokines with type 2 diabetes mellitus and diabetic nephropathy: a bidirectional Mendelian randomization study.
Song S, Ni J, Sun Y, Pu Q, Zhang L, Yan Q, Yu J. Song S, et al. Front Med (Lausanne). 2024 Nov 7;11:1459752. doi: 10.3389/fmed.2024.1459752. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39574905 Free PMC article. - Deciphering molecular landscape of breast cancer progression and insights from functional genomics and therapeutic explorations followed by in vitro validation.
Khan B, Qahwaji R, Alfaifi MS, Athar T, Khan A, Mobashir M, Ashankyty I, Imtiyaz K, Alahmadi A, Rizvi MMA. Khan B, et al. Sci Rep. 2024 Nov 20;14(1):28794. doi: 10.1038/s41598-024-80455-6. Sci Rep. 2024. PMID: 39567714 Free PMC article. - Quantitative DCE Dynamics on Transformed MR Imaging Discriminates Clinically Significant Prostate Cancer.
Wei Z, Iluppangama M, Qi J, Choi JW, Yu A, Gage K, Chumbalkar V, Dhilon J, Balaji KC, Venkataperumal S, Hernandez DJ, Park J, Yedjou C, Alo R, Gatenby RA, Pow-Sang J, Balagurunanthan Y. Wei Z, et al. Cancer Control. 2024 Jan-Dec;31:10732748241298539. doi: 10.1177/10732748241298539. Cancer Control. 2024. PMID: 39545376 Free PMC article. - Analysis of behavioral flow resolves latent phenotypes.
von Ziegler LM, Roessler FK, Sturman O, Waag R, Privitera M, Duss SN, O'Connor EC, Bohacek J. von Ziegler LM, et al. Nat Methods. 2024 Dec;21(12):2376-2387. doi: 10.1038/s41592-024-02500-6. Epub 2024 Nov 12. Nat Methods. 2024. PMID: 39533008 Free PMC article.
References
- Lander, E. S. & Kruglyak, L. (1995) Nat. Genet. 11, 241–247. - PubMed
- Storey, J. D. (2003) Ann. Stat., in press.
- Storey, J. D. (2002) J. R. Stat. Soc. B 64, 479–498.
- Benjamini, Y. & Hochberg, Y. (1995) J. R. Stat. Soc. B 85, 289–300.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources