Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles - PubMed (original) (raw)
Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles
Ana Paula V Castro et al. J Virol. 2003 Aug.
Abstract
Cyclophilins are peptidyl-prolyl cis-trans isomerases involved in catalyzing conformational changes and accelerating the rate of protein folding and refolding in several cellular systems. In the present study, we analyzed the expression pattern and intracellular distribution of the cellular isomerase cyclophilin A (CypA) during vaccinia virus (VV) infection. An impressive increase in CypA stability was observed, leading to a practically unchanged accumulation of CypA during infection, although its synthesis was completely inhibited at late times. By confocal microscopy, we observed that CypA went through an intense reorganization in the cell cytoplasm and colocalized with the virosomes late in infection. CypA relocation to viral factories required the synthesis of viral postreplicative proteins, and treatment of infected cells with cyclosporine (CsA) prevented CypA relocation, clearly excluding the virosomes from CypA staining. Immunoelectron microscopy of VV-infected cells showed that CypA was incorporated into VV particles during morphogenesis. Biochemical and electron microscopic assays with purified virions confirmed that CypA was encapsidated within the virus particle and localized specifically in the core. This work suggests that CypA may develop an important role in VV replication.
Figures
FIG. 1.
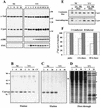
Temporal synthesis and accumulation of CypA in VV-infected cells. BSC-40 cells were mock infected (NI) or infected with VV at 10 PFU/ml, and at the indicated times p.i., postnuclear cell extracts were prepared. (A) Proteins were resolved by SDS-13.5% PAGE, followed by Western blot analysis with anti-α-tubulin, anti-CypA, anti-F11L, and anti-A18R antibodies. (B, C, and D) At the indicated times p.i., the monolayers were pulse-labeled with [35S]methionine for 1 h, extracts were prepared, and 200-μg samples were loaded onto 8′A-Cs-Affi-Gel columns. The proteins retained by the resin were eluted with 2 mg of CsA per ml. Both the unbound (flowthrough) and eluted proteins were analyzed by SDS-13.5% PAGE and staining with Coomassie blue (B), followed by autoradiography (C and D). Asterisks indicate late VV proteins; H indicates a major host protein. CT refers to the control column unable to retain CypA. (E) At 2 h p.i., the monolayers were labeled with [35S]methionine (pulse; P) as described above and chased with medium containing unlabeled methionine for an additional 6 and 18 h. The extracts were prepared and loaded onto 8′A-Cs-Affi-Gel 10 columns as already described. The eluted proteins were resolved by SDS-PAGE and staining with Coomassie blue, followed by autoradiography. (F) The values obtained by densitometric analysis of the autoradiograms in panel E were corrected by the amount of eluted CypA obtained for the corresponding lane in the Coomassie blue-stained gels. The resulting ratio was calculated as described in Materials and Methods, and the final values are expressed as percentages of the 35S-labeled CypA synthesized at the time of the pulse (baseline). The values to the right of panel A and the left of panel B are molecular sizes in kilodaltons.
FIG. 2.
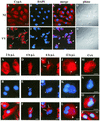
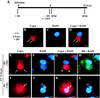
(Top panels) Intracellular localization of CypA in VV-infected cells. BSC-40 cells were mock infected (NI) or infected with VV at 10 PFU/ml. At 20 h p.i., the cells were processed for confocal immunofluorescence microscopy with anti-CypA detected with a Cy3-conjugated secondary antibody (red channel; A and E). Viral and cellular DNAs were stained with DAPI (B and F). The merged images are shown in panels C and G. Phase-contrast images are shown in panels D and H. Arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Arrowheads point to colocalization of CypA with viral factories in the merged images. Representative fields are shown. N, nucleus; bar, 20 μm.
FIG. 3.
(Bottom panels) Time course of CypA relocalization. BSC-40 cells were mock infected or infected with VV as described in Materials and Methods and harvested for immunofluorescence assay at the indicated times (A to L). In panels M to P, the cells were mock infected (P) or infected with VV (M, N, and O) and treated with 35 μM CsA at time zero. The infection proceeded for 12 h p.i., when the cells were processed for immunofluorescence assay. Anti-CypA labeling (red channel) is visualized in panels A, D, G, J, and M. Viral and cellular DNAs were stained with DAPI (blue channel), as shown in panels B, E, H, K, and N. Merged images are visualized in panels C, F, I, L, O, and P. White arrows indicate granular CypA structures in the cytoplasm. Thin arrows indicate the redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Arrowheads point to colocalization of CypA with viral factories in the merged images. Open arrows indicate CypA labeling outlining viral factories. Representative fields are shown. NI, mock-infected cells; N, nucleus; bars, 10 μm.
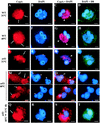
FIG. 4.
Distribution of CypA and α-tubulin in VV-infected cells. BSC-40 cells (A to E) and mouse embryo fibroblasts (F to H) were mock infected (A, B, and F) or infected with VV (C, D, E, G, and H) as described in Materials and Methods. At 20 h p.i., the monolayers were processed for immunofluorescence microscopy with anti-α-tubulin (A and C) and anti-CypA (B, D, F, and G) antibodies. Viral and cellular DNAs were stained with DAPI (E and H). Arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Representative fields are shown. N, nucleus; bars, 10 μm.
FIG. 5.
Effects of metabolic inhibitors on the distribution of CypA. BSC-40 cells infected with VV were treated with 40 μg of AraC per ml for 10 h (A and B) or with 100 μg of CHX per ml for 6 h (E and F). Cells were processed for immunofluorescence microscopy with anti-CypA (A, C, and E) antibody. Viral and cellular DNAs were stained with DAPI (B, D, and F). (C and D) Untreated cells infected for 10 h. Thin arrows indicate granular CypA structures in the cytoplasm. White arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Representative fields are shown. N, nucleus; bars, 10 μm.
FIG. 6.
Localization of CypA in VV-infected cells in the absence of postreplicative protein synthesis. (A) Diagram of the assay. BSC-40 cells were infected with VV in the presence of 10 mM HU. At 6 h p.i. (B to D), the medium was replaced with HU-free medium in the absence (E to H) or presence (I to L) of 100 μg of CHX per ml. The cells were incubated for an additional 6 h and processed for immunofluorescence (IFM) with anti-CypA (B, E, and I) and anti-D8L (green channel; H and L) antibodies. Viral and cellular DNAs were stained with DAPI (C, F, and J). Merged images are shown in panels D, G, K, H, and L. Thin arrows indicate granular CypA structures in the cytoplasm. White arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Asterisks indicate viral factories. Arrowheads point to colocalization of CypA with viral factories. Representative fields are shown. N, nucleus; bars, 10 μm.
FIG. 7.
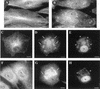
Localization of CypA in _ts_53-infected cells. BSC-40 cells were infected with WT VV (A to H) or the VV mutant _ts_53 (I to T) at 31 or 40°C. After 12 h, the cells were processed for immunofluorescence microscopy with anti-CypA (A, E, I, M, and Q) and anti-D8L (green channel; D, H, L, P, and T) antibodies. Viral and cellular DNAs were stained with DAPI (B, F, J, N, and R). Merged images are shown in panels C, D, G, H, K, L, O, P, S, and T. In panels Q to T, _ts_53-infected cells were incubated for 12 h at 40°C and shifted to 31°C for 4 h, after which they were processed for immunofluorescence assay. White arrows indicate redistribution of CypA to perinuclear regions of VV-infected cells. Open arrows point to infected cells with no redistribution of CypA. Thin arrows point to redistribution of CypA to the periphery of the virosomes. Asterisks indicate viral factories. White arrowheads indicate colocalization of CypA with viral factories. Open arrowheads indicate the absence of colocalization of CypA with virosomes. Representative fields are shown. N, nucleus; bars, 10 μm.
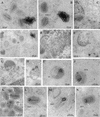
FIG. 8.
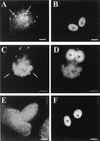
Localization of CypA in VV-infected cells by immunoelectron microscopy. BSC-40 cells mock infected (C) or infected with VV for 20 h were fixed and processed for immunogold labeling with anti-CypA antibody, followed by a colloidal gold-conjugated secondary antibody. Representative fields are shown. CypA labeling is indicated by arrows. In panels A, B, and D, note the intense labeling of amorphous electron-dense material in the proximity of VV particles. Arrowheads in panel E point to gold labeling in the periphery and within the viroplasm. In panels F to H, note the association of gold grains with the viroplasm being encapsidated by the crescent forms. CypA labeling is also found associated with electron-dense material and nucleoid of IVs (I and J). In mature particles, gold labeling is primarily found in the core (K to N). Cr, crescent forms; IVn, IVs with nucleoid; IEV, intracellular enveloped virus; N, nucleus.
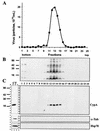
FIG. 9.
Association of CypA with purified VV. Intracellular virus was sedimented on a sucrose cushion and banded on three successive sucrose gradients. Fractions were collected, and the number of particles was determined by OD measurement (A). Aliquots of each fraction were analyzed by SDS-PAGE, followed by a Western blot assay with antiserum raised against total VV proteins (B), anti-CypA (C), anti-α-tubulin, and anti-Hsp70 (C) antibodies. In lane CT, uninfected BSC-40 cell proteins were analyzed by Western blotting as a positive control for the anti-CypA, anti-α-tubulin, and anti-Hsp70 antibodies. Molecular size markers (kilodaltons) are shown to the left of panels B and C.
FIG. 10.
Association of CypA with VV cores. (A and B) Purified VV (7.5 × 109 particles) was incubated with 0.5% NP-40 in the presence or absence of 50 mM DTT. The insoluble core fraction was recovered by centrifugation and treated with 0.2% deoxycholate (DOC). Insoluble and soluble fractions were obtained by centrifugation. The proteins were resolved by SDS-PAGE, followed by silver staining (A). The samples were also analyzed by Western blotting (B) with anti-CypA, anti-A18R, anti-D12R, anti-D1R, anti-A12L, and anti-H3L antibodies. T refers to untreated VV. I and S refer to insoluble and soluble fractions, respectively. (C) Purified VV (7.5 × 109 particles) was digested with different concentrations of proteinase K or with 50 μg of chymotrypsin per ml. The proteins in each sample were resolved by SDS-PAGE, followed by Western blot analysis with anti-CypA, anti-A18R, anti-D12R, anti-D8L, and anti-H3L antibodies. The values to the left of panel A are molecular sizes in kilodaltons.
FIG. 11.
Detection of CypA in purified VV by immunogold labeling and negative staining. Purified VV (1.5 × 108 particles) was adsorbed to Formvar-coated nickel grids and left unpermeabilized (A, C, E, and F) or permeabilized with Triton X-100 (B and G) or NP-40-DTT (D). The grids were incubated separately with anti-CypA (A to D), anti-D8L, anti-H3L (E), or anti-D12R (F and G) antibody, followed by a colloidal gold-conjugated secondary antibody. Representative fields are shown.
Similar articles
- Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype.
Bose S, Mathur M, Bates P, Joshi N, Banerjee AK. Bose S, et al. J Gen Virol. 2003 Jul;84(Pt 7):1687-1699. doi: 10.1099/vir.0.19074-0. J Gen Virol. 2003. PMID: 12810862 - Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication.
Liu X, Sun L, Yu M, Wang Z, Xu C, Xue Q, Zhang K, Ye X, Kitamura Y, Liu W. Liu X, et al. Cell Microbiol. 2009 May;11(5):730-41. doi: 10.1111/j.1462-5822.2009.01286.x. Epub 2009 Jan 15. Cell Microbiol. 2009. PMID: 19207730 - trans processing of vaccinia virus core proteins.
Lee P, Hruby DE. Lee P, et al. J Virol. 1993 Jul;67(7):4252-63. doi: 10.1128/JVI.67.7.4252-4263.1993. J Virol. 1993. PMID: 7685413 Free PMC article. - Insights into the roles of cyclophilin A during influenza virus infection.
Liu X, Zhao Z, Liu W. Liu X, et al. Viruses. 2013 Jan 15;5(1):182-91. doi: 10.3390/v5010182. Viruses. 2013. PMID: 23322171 Free PMC article. Review. - Cyclophilin A: A Key Factor in Virus Replication and Potential Target for Anti-viral Therapy.
Dawar FU, Tu J, Khattak MN, Mei J, Lin L. Dawar FU, et al. Curr Issues Mol Biol. 2017;21:1-20. doi: 10.21775/cimb.021.001. Epub 2016 Mar 31. Curr Issues Mol Biol. 2017. PMID: 27033630 Review.
Cited by
- CD147/EMMPRIN acts as a functional entry receptor for measles virus on epithelial cells.
Watanabe A, Yoneda M, Ikeda F, Terao-Muto Y, Sato H, Kai C. Watanabe A, et al. J Virol. 2010 May;84(9):4183-93. doi: 10.1128/JVI.02168-09. Epub 2010 Feb 10. J Virol. 2010. PMID: 20147391 Free PMC article. - Independent evolution of an antiviral TRIMCyp in rhesus macaques.
Wilson SJ, Webb BL, Ylinen LM, Verschoor E, Heeney JL, Towers GJ. Wilson SJ, et al. Proc Natl Acad Sci U S A. 2008 Mar 4;105(9):3557-62. doi: 10.1073/pnas.0709003105. Epub 2008 Feb 19. Proc Natl Acad Sci U S A. 2008. PMID: 18287035 Free PMC article. - A multidisciplinary approach to study a couple of monozygotic twins discordant for the chronic fatigue syndrome: a focus on potential salivary biomarkers.
Ciregia F, Giusti L, Da Valle Y, Donadio E, Consensi A, Giacomelli C, Sernissi F, Scarpellini P, Maggi F, Lucacchini A, Bazzichi L. Ciregia F, et al. J Transl Med. 2013 Oct 2;11:243. doi: 10.1186/1479-5876-11-243. J Transl Med. 2013. PMID: 24088505 Free PMC article. - TRIM5α restricts poxviruses and is antagonized by CypA and the viral protein C6.
Zhao Y, Lu Y, Richardson S, Sreekumar M, Albarnaz JD, Smith GL. Zhao Y, et al. Nature. 2023 Aug;620(7975):873-880. doi: 10.1038/s41586-023-06401-0. Epub 2023 Aug 9. Nature. 2023. PMID: 37558876 Free PMC article. - Vaccinia virus mutations in the L4R gene encoding a virion structural protein produce abnormal mature particles lacking a nucleocapsid.
Jesus DM, Moussatche N, Condit RC. Jesus DM, et al. J Virol. 2014 Dec;88(24):14017-29. doi: 10.1128/JVI.02126-14. Epub 2014 Sep 24. J Virol. 2014. PMID: 25253347 Free PMC article.
References
- Beaud, G., and R. Beaud. 1997. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J. Gen. Virol. 78:3297-3302. - PubMed
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
- Bristow, R., J. Byrne, J. Squirell, H. Trencher, T. Carter, B. Rodgers, E. Saman, and J. Duncan. 1999. Human cyclophilin has a significantly higher affinity for HIV-1 recombinant p55 than p24. J. Acquir. Immune Defic. Syndr. Hum. Retrovir. 20:334-336. - PubMed
- Brown, C. R., D. Y. Cui, G. G. Hung, and H. L. Chiang. 2001. Cyclophilin A mediates Vid22p function in the import of fructose-1,6-bisphosphatase into Vid vesicles. J. Biol. Chem. 276:48017-48026. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous