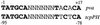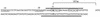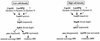The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome - PubMed (original) (raw)
The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome
Gabriela Kovacikova et al. J Bacteriol. 2003 Aug.
Abstract
Activation of the tcpPH promoter on the Vibrio pathogenicity island by AphA and AphB initiates the Vibrio cholerae virulence cascade and is regulated by quorum sensing through the repressive action of HapR on aphA expression. To further understand how the chromosomally encoded AphA protein activates tcpPH expression, site-directed mutagenesis was used to identify the base pairs critical for AphA binding and transcriptional activation. This analysis revealed a region of partial dyad symmetry, TATGCA-N6-TNCNNA, that is important for both of these activities. Searching the V. cholerae genome for this binding site permitted the identification of a second one upstream of a penicillin V amidase (PVA) gene on the small chromosome. AphA binds to and footprints this site, which overlaps the pva transcriptional start, consistent with its role as a repressor at this promoter. Since aphA expression is under quorum-sensing control, the response regulators LuxO and HapR also influence pva expression. Thus, pva is repressed at low cell density when AphA levels are high, and it is derepressed at high cell density when AphA levels are reduced. Penicillin amidases are thought to function as scavengers for phenylacetylated compounds in the nonparasitic environment. That AphA oppositely regulates the expression of pva from that of virulence, together with the observation that PVA does not play a role in virulence, suggests that these activities are coordinated to serve V. cholerae in different biological niches.
Figures
FIG. 1.
Schematic showing the positions of the primers used to introduce the AphA binding site changes into the tcpPH promoter. The base pairs that were individually changed, from −75 to −98, are shown in bold. wt, wild type.
FIG. 2.
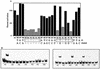
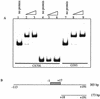
Mutational analysis of the AphA binding site at the tcpPH promoter. (A) V. cholerae strains were grown in LB (pH 6.5) at 30°C. From left to right: KSK618 (wild-type [wt] tcpP-lacZ); WL56 (−98A), WL55 (−97C), WL52 (−96A), WL57 (−95A), WL38 (−94C), WL39 (−93A), WL37 (−92T), WL36 (−91G), WL35 (−90C), WL40 (−89C), GK873 (−88A), GK872 (−87G), GK848 (−86C), GK847 (−85C), GK846 (−84T), GK845 (−83A), GK801 (−82G), GK835 (−81A), GK834 (−80G), KSK1309 (−79G), GK800 (−78C), GK799 (−77G), GK798 (−76A), and GK797 (−75C). Positions reducing expression of the promoter to 34% or lower are shaded in gray, and the corresponding bases are in gray. (B) AphA binding to various 135-bp mutant promoter fragments from −175 to −40. The first lane in each set has no protein added, and the second lane has 5 ng (15 nM) of AphA added.
FIG. 3.
Comparison of the AphA binding sites at the tcpPH and pva promoters. Conserved positions are shown in bold.
FIG. 4.
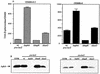
Influence of Δ_aphA_, Δ_hapR_, and Δ_luxO_ mutations on a C6706 pva_-lacZ fusion at low and high cell density. Strains were grown in AKI conditions for 3.5 h (OD600 = 0.2) or for 7.5 h (OD600 = 4). (A) From left to right: GK925 (wild-type [wt] pva-lacZ); GK927 (ΔaphA); KSK1918 (Δ_hapR); KSK1920 (Δ_luxO_). (B) Anti-AphA Western blot with strains grown as described for panel A. From left to right: C6706, GK925, GK927, KSK1918, and KSK1920.
FIG. 5.
Nucleotide sequence of the proximal region of the pva promoter. The transcriptional start site (+1), ATG codon, and −10 and −35 regions are shown. Boxes show the region of dyad symmetry involved in AphA recognition, and thick lines show the base pairs that are protected from DNaseI digestion by AphA.
FIG. 6.
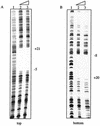
A. Binding of purified AphA to the pva promoter. Lanes 1 to 3, a 303-bp fragment amplified from C6076; lanes 4 to 6, a 173-bp fragment amplified from C6706; lanes 7 to 9, the 303-bp fragment amplified from O395. The first lane in each set has no protein added, the second lane has 5 ng (15 nM) of AphA, and the third lane has 50 ng (150 nM) of AphA. (B) Diagrammatic representation of the binding fragments used for panel A. The gray box shows the AphA binding site.
FIG. 7.
DNaseI footprint for AphA at the pva promoter. (A) Top strand of a 233-bp fragment from −113 to +120. Lane 1, no protein; lane 2, 250 ng (2.5 μM) of AphA; lane 3, 300 ng (3 μM) of AphA. (B) Bottom strand of the 233-bp fragment. Lanes 1 to 3 are the same as those described for panel A.
FIG. 8.
Contrasting pathways of gene expression regulated by AphA in response to cell density. At least three sensory circuits, CqsAS (System 1), LuxSPQ (System 2), and the unknown components of proposed System 3 function in parallel to control the activity of LuxO in response to the presence of autoinducers. At low cell density, activated LuxO indirectly represses the expression of hapR. The resulting high levels of AphA repress the expression of pva and, in the presence of AphB, activate tcpPH expression and the rest of the virulence cascade. At high cell density, inactive LuxO no longer represses hapR expression. HapR then reduces AphA levels by repressing expression from the aphA promoter. This derepresses pva expression while simultaneously preventing the activation of tcpPH and the rest of the virulence cascade.
Similar articles
- Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter.
Kovacikova G, Skorupski K. Kovacikova G, et al. Mol Microbiol. 2001 Jul;41(2):393-407. doi: 10.1046/j.1365-2958.2001.02518.x. Mol Microbiol. 2001. PMID: 11489126 - Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter.
Kovacikova G, Skorupski K. Kovacikova G, et al. Mol Microbiol. 2002 Nov;46(4):1135-47. doi: 10.1046/j.1365-2958.2002.03229.x. Mol Microbiol. 2002. PMID: 12421317 - Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter.
Kovacikova G, Lin W, Skorupski K. Kovacikova G, et al. Mol Microbiol. 2004 Jul;53(1):129-42. doi: 10.1111/j.1365-2958.2004.04121.x. Mol Microbiol. 2004. PMID: 15225309 - Regulation of virulence in Vibrio cholerae.
Klose KE. Klose KE. Int J Med Microbiol. 2001 May;291(2):81-8. doi: 10.1078/1438-4221-00104. Int J Med Microbiol. 2001. PMID: 11437342 Review.
Cited by
- PhoB regulates both environmental and virulence gene expression in Vibrio cholerae.
Pratt JT, Ismail AM, Camilli A. Pratt JT, et al. Mol Microbiol. 2010 Sep;77(6):1595-605. doi: 10.1111/j.1365-2958.2010.07310.x. Epub 2010 Aug 16. Mol Microbiol. 2010. PMID: 20659293 Free PMC article. - Virulence regulator AphB enhances toxR transcription in Vibrio cholerae.
Xu X, Stern AM, Liu Z, Kan B, Zhu J. Xu X, et al. BMC Microbiol. 2010 Jan 6;10:3. doi: 10.1186/1471-2180-10-3. BMC Microbiol. 2010. PMID: 20053280 Free PMC article. - Regulatory Hierarchies Controlling Virulence Gene Expression in Shigella flexneri and Vibrio cholerae.
Dorman MJ, Dorman CJ. Dorman MJ, et al. Front Microbiol. 2018 Nov 9;9:2686. doi: 10.3389/fmicb.2018.02686. eCollection 2018. Front Microbiol. 2018. PMID: 30473684 Free PMC article. Review. - Calcium Enhances Bile Salt-Dependent Virulence Activation in Vibrio cholerae.
Hay AJ, Yang M, Xia X, Liu Z, Hammons J, Fenical W, Zhu J. Hay AJ, et al. Infect Immun. 2016 Dec 29;85(1):e00707-16. doi: 10.1128/IAI.00707-16. Print 2017 Jan. Infect Immun. 2016. PMID: 27849180 Free PMC article. - Molecular characterization of direct target genes and cis-acting consensus recognized by quorum-sensing regulator AphA in Vibrio parahaemolyticus.
Sun F, Zhang Y, Wang L, Yan X, Tan Y, Guo Z, Qiu J, Yang R, Xia P, Zhou D. Sun F, et al. PLoS One. 2012;7(9):e44210. doi: 10.1371/journal.pone.0044210. Epub 2012 Sep 12. PLoS One. 2012. PMID: 22984476 Free PMC article.
References
- Carroll, P. A., K. T. Tashima, M. B. Rogers, V. J. DiRita, and S. B. Calderwood. 1997. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 25:1099-1111. - PubMed
- Champion, G. A., M. N. Neely, M. A. Brennan, and V. J. DiRita. 1997. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol. Microbiol. 23:323-331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous