Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27 - PubMed (original) (raw)
Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27
Jinrong Min et al. Genes Dev. 2003.
Abstract
The chromodomain of Drosophila Polycomb protein is essential for maintaining the silencing state of homeotic genes during development. Recent studies suggest that Polycomb mediates the assembly of repressive higher-order chromatin structures in conjunction with the methylation of Lys 27 of histone H3 by a Polycomb group repressor complex. A similar mechanism in heterochromatin assembly is mediated by HP1, a chromodomain protein that binds to histone H3 methylated at Lys 9. To understand the molecular mechanism of the methyl-Lys 27 histone code recognition, we have determined a 1.4-A-resolution structure of the chromodomain of Polycomb in complex with a histone H3 peptide trimethylated at Lys 27. The structure reveals a conserved mode of methyl-lysine binding and identifies Polycomb-specific interactions with histone H3. The structure also reveals a dPC dimer in the crystal lattice that is mediated by residues specifically conserved in the Polycomb family of chromodomains. The dimerization of dPC can effectively account for the histone-binding specificity and provides new mechanistic insights into the function of Polycomb. We propose that self-association is functionally important for Polycomb.
Figures
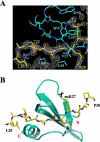
Figure 1.
Structure of the dPC chromodomain in complex with the H3m3K27 peptide. (A) A simulated annealing omit electron density map showing the binding of H3m3K27. The 1.4 Å (FO - FC, ϕC) difference map was calculated with the peptide omitted, and the map is contoured at the 1.0σ contour level with a 3.0σ cutoff. The refined structure is superimposed as a bond model: red, oxygen; blue, nitrogen; cyan in dPC, carbon; yellow, carbon in the peptide. Key residues of histone H3 and dPC are labeled as follows: cyan, dPC; yellow, histone H3. (B) Overall structure of the dPC chromodomain-H3m3K27 peptide complex. dPC is shown in a ribbon representation, and the H3m3K27 peptide is shown as a bond model.
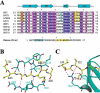
Figure 2.
Chromodomain-histone interactions. (A) Alignment of chromodomain sequences of Drosophila and human PC and HP1 proteins. dPC,Drosophila Polycomb (SWISS-PROT database accession code P26017); hPC2, human Polycomb 2 homolog (SWISS-PROT O00257); hCBX6, human chromobox homolog 6 (SWISS-PROT O95503); hPC3, human Polycomb 3 homolog (SWISSPROT Q9HC52); dHP1, Drosophila HP1 (SWISS-PROT P05205). hHP1α, hHP1β, and hHP1γ are human HP1 homologs with the SWISS-PROT accession codes P45973, P23197, and Q13185, respectively. Identical residues among these proteins are shown in white letters over a blue background, and similar residues are shown in white letters over a purple background. Amino acids specifically conserved in the PC family are highlighted: Yellow indicates residues involved in PC-specific interactions with histone H3, and the residues enclosed in a red box are involved in the formation of a dPC dimer. Tyr 54 highlighted in cyan is located at the methyl-lysine binding pocket. Every 10 residues are indicated with a “+” above the sequence. Secondary structural elements and their nomenclatures are shown_above_ the sequences. The sequence of the N-terminal tail of histone H3 is also shown. HP1-interacting residues are highlighted in cyan, and the residues interacting with the chromodomain of dPC are highlighted in yellow. Residues shown in red letters are important for specific binding to dPC. (B) A bond model showing extensive hydrogen bonding between main-chain atoms of dPC and the H3m3K27 peptide. Hydrogen bonds are drawn in magenta broken lines, and the same color code as in Figure 1 is used for dPC and H3m3K27. (C) Unique interactions between dPC and H3m3K27 occur at the N-terminal end of the peptide. The H3m3K27 peptide and Arg 67 and Asp 65 of dPC are shown in a bond model superimposed with the ribbon diagram of dPC.
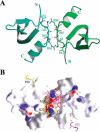
Figure 3.
The dPC chromodomain dimer. (A) Two dPC chromodomain monomers are shown in a ribbon representation, colored cyan and green, respectively. Key residues involved in dimerization are shown in a bond model. Hydrogen bonds involving these residues are indicated with magenta broken lines. (B) The dPC chromodomain dimer juxtaposes the two binding sites of H3m3K27. The dPC chromodomain dimer is shown in a surface representation. The dimer is viewed from a similar direction as in A. Surface areas colored blue or red indicate positive or negative electric potentials. Two bound H3m3K27 peptides are shown in a bond model. Carbon atoms in the second peptide are colored magenta.
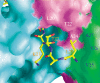
Figure 4.
A shallow hydrophobic binding pocket of the N-terminal region of one H3m3K27 peptide. Two dPC molecules and a second H3m3K27 are shown in a surface representation. Cyan and green surfaces are that of the two dPC molecules (same coloring scheme as in Fig. 3A), and the magenta surface is that of another H3m3K27 peptide. The binding site is formed by Val 25, Tyr 26, and Ala 27 of the other dPC chromodomain (green surface; residues labeled in blue letters) and N-terminal residues of a separate H3m3K27 peptide (magenta surface; residues labeled in white letters) in the dimer.
Similar articles
- Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains.
Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Fischle W, et al. Genes Dev. 2003 Aug 1;17(15):1870-81. doi: 10.1101/gad.1110503. Genes Dev. 2003. PMID: 12897054 Free PMC article. - Double chromodomains cooperate to recognize the methylated histone H3 tail.
Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Flanagan JF, et al. Nature. 2005 Dec 22;438(7071):1181-5. doi: 10.1038/nature04290. Nature. 2005. PMID: 16372014 - Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Li J, Li Z, Ruan J, Xu C, Tong Y, Pan PW, Tempel W, Crombet L, Min J, Zang J. Li J, et al. PLoS One. 2011;6(10):e25104. doi: 10.1371/journal.pone.0025104. Epub 2011 Oct 12. PLoS One. 2011. PMID: 22022377 Free PMC article. - The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3.
Cao R, Zhang Y. Cao R, et al. Curr Opin Genet Dev. 2004 Apr;14(2):155-64. doi: 10.1016/j.gde.2004.02.001. Curr Opin Genet Dev. 2004. PMID: 15196462 Review. - Phosphorylation of repressive histone code readers by casein kinase 2 plays diverse roles in heterochromatin regulation.
Murakami Y. Murakami Y. J Biochem. 2019 Jul 1;166(1):3-6. doi: 10.1093/jb/mvz045. J Biochem. 2019. PMID: 31198932 Review.
Cited by
- Epigenetic regulation of skin: focus on the Polycomb complex.
Zhang J, Bardot ES, Ezhkova E. Zhang J, et al. Cell Mol Life Sci. 2012 Jul;69(13):2161-2172. doi: 10.1007/s00018-012-0920-x. Cell Mol Life Sci. 2012. PMID: 22314499 Free PMC article. Review. - Histone Readers and Their Roles in Cancer.
Wen H, Shi X. Wen H, et al. Cancer Treat Res. 2023;190:245-272. doi: 10.1007/978-3-031-45654-1_8. Cancer Treat Res. 2023. PMID: 38113004 Free PMC article. - A novel human polycomb binding site acts as a functional polycomb response element in Drosophila.
Cuddapah S, Roh TY, Cui K, Jose CC, Fuller MT, Zhao K, Chen X. Cuddapah S, et al. PLoS One. 2012;7(5):e36365. doi: 10.1371/journal.pone.0036365. Epub 2012 May 3. PLoS One. 2012. PMID: 22570707 Free PMC article. - Tudor, MBT and chromo domains gauge the degree of lysine methylation.
Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Kim J, et al. EMBO Rep. 2006 Apr;7(4):397-403. doi: 10.1038/sj.embor.7400625. Epub 2006 Jan 13. EMBO Rep. 2006. PMID: 16415788 Free PMC article. - In vivo analysis of Drosophila SU(Z)12 function.
Chen S, Birve A, Rasmuson-Lestander A. Chen S, et al. Mol Genet Genomics. 2008 Feb;279(2):159-70. doi: 10.1007/s00438-007-0304-3. Epub 2007 Nov 22. Mol Genet Genomics. 2008. PMID: 18034266
References
- Bannister A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120-124. - PubMed
- Brasher S.V., Smith, B.O., Fogh, R.H., Nietlispach, D., Thiru, A., Nielsen, P.R., Broadhurst, R.W., Ball, L.J., Murzina, N.V., and Laue, E.D. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19: 1587-1597. - PMC - PubMed
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54: 905-921. - PubMed
- Cao R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., Jones, R.S., and Zhang, Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039-1043. - PubMed
- Cowell I.G. and Austin, C.A. 1997. Self-association of chromo domain peptides. Biochim. Biophys. Acta 1337: 198-206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases