Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b - PubMed (original) (raw)
Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b
Taiping Chen et al. Mol Cell Biol. 2003 Aug.
Abstract
We have previously shown that the DNA methyltransferases Dnmt3a and Dnmt3b carry out de novo methylation of the mouse genome during early postimplantation development and of maternally imprinted genes in the oocyte. In the present study, we demonstrate that Dnmt3a and Dnmt3b are also essential for the stable inheritance, or "maintenance," of DNA methylation patterns. Inactivation of both Dnmt3a and Dnmt3b in embryonic stem (ES) cells results in progressive loss of methylation in various repeats and single-copy genes. Interestingly, introduction of the Dnmt3a, Dnmt3a2, and Dnmt3b1 isoforms back into highly demethylated mutant ES cells restores genomic methylation patterns; these isoforms appear to have both common and distinct DNA targets, but they all fail to restore the maternal methylation imprints. In contrast, overexpression of Dnmt1 and Dnmt3b3 failed to restore DNA methylation patterns due to their inability to catalyze de novo methylation in vivo. We also show that hypermethylation of genomic DNA by Dnmt3a and Dnmt3b is necessary for ES cells to form teratomas in nude mice. These results indicate that genomic methylation patterns are determined partly through differential expression of different Dnmt3a and Dnmt3b isoforms.
Figures
FIG. 1.
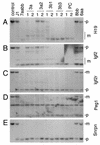
Inactivation of Dnmt3a and Dnmt3b results in progressive loss of DNA methylation in ES cells. (A) Genomic DNA from Dnmt3a−/− Dnmt3b−/− ES cells (7aabb and 10aabb) that had been grown in culture for 5 to 40 passages, as well as wild-type (J1) and Dnmt1 mutant (n/n and c/c) ES cells, was digested with _Hpa_II and hybridized to probes for endogenous C-type retrovirus repeats (pMO), minor satellite repeats, and IAP repeats. As a control for complete digestion, DNA from J1 cells was digested with _Msp_I. The Dnmt1n allele (the “n” stands for N-terminal disruption) is a partial loss-of-function mutation (27) and the Dnmt1c allele (the “c” stands for disruption of the catalytic or C-terminal domain) is a null mutation (24). (B) Genomic DNA from J1, Dnmt3a−/− (6aa), or Dnmt3b−/− (8bb) ES cells that had been grown in culture for 5 to 25 passages, as well as 7aabb (P40), was digested with _Hpa_II and hybridized to the pMO probe. (C) Lysates from the indicated ES cell lines were immunoblotted with anti-Dnmt1 and anti-tubulin antibodies.
FIG. 2.
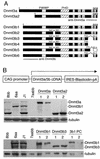
Stable expression of Dnmt3a and Dnmt3b isoforms in late-passage 7aabb cells. (A) Schematic diagram of Dnmt3a and Dnmt3b isoforms. The conserved PWWP and PHD domains, the methyltransferase motifs (I, IV, VI, IX, and X), and the sites of alternative splicing are indicated (the C-terminal 45 amino acids of Dnmt3b5 are out of frame and shown as an open bar). The locations of the epitopes for the Dnmt3a and Dnmt3b antibodies are also shown. (B) cDNAs encoding Dnmt3a/3b isoforms were subcloned in an expression vector (schematically shown at the top), and these constructs were individually electroporated into late-passage (P70) 7aabb cells, which were subsequently selected in blasticidin-containing medium for 7 days. Blasticidin-resistant clones were analyzed with immunoblotting with anti-Dnmt3a (middle panel) or anti-Dnmt3b (bottom panel) antibodies. As a loading control, the same membranes were immunoblotted with anti-tubulin antibody.
FIG. 3.
Expression of Dnmt3a/3b proteins in 7aabb cells restores DNA methylation. (A to D) Methylation of repetitive sequences. Genomic DNA from the indicated ES cell lines was digested with _Hpa_II (A to C) or _Mae_II (D) and hybridized to the indicated probes. DNA from J1 cells digested with _Msp_I was used as a control for complete digestion. (E) Analysis of the methylation status of the major satellite repeating unit by bisulfite sequencing. Genomic DNA from J1 and 7aabb cells, as well as from stable cell lines expressing Dnmt3a, Dnmt3a2, Dnmt3b1, and Dnmt3b3, was analyzed. The methylation status of six CpG sites from 8 to 12 individual clones is shown schematically (black circles represent methylated sites), and the percentages of methylated CpG sites are indicated in parenthesis. (F to I) Methylation of unique genes. The genomic DNA samples described in panels A to D were digested with _Bam_HI and _Hha_I (F and H), _Eco_RI and _Hpa_II (G), or _Eco_RV and Hha_I (I) and hybridized to probes corresponding to the 3′ region of β_-globin (F), the 5′ region of Pgk-1 (G), an exon of Pgk-2 (H), or the 5′ region of Xist (I). DNA from J1 cells digested with _Bam_HI alone (F and H) or _Eco_RI alone (G) was used as controls.
FIG. 4.
Expression of Dnmt3a and Dnmt3b proteins in 7aabb cells fails to restore maternal methylation imprints. The same DNA samples described in Fig. 3 were digested with _Sac_I and _Hha_I (A), _Bam_HI and _Hpa_II (B), _Pvu_II and _Hpa_II (C and D), or _Xba_I and _Hha_I (E) and hybridized to probes corresponding to the 5′ upstream region of H19 (A), the DMR2 of Igf2 (B), region 2 of Igf2r (C), the DMR of Peg1 (D), or the DMR1 of Snrpn (E). As controls, DNA from J1 cells was digested with the corresponding enzymes without _Hha_I or _Hpa_II. The fragments derived from the paternal (p) and maternal (m) alleles are indicated.
FIG. 5.
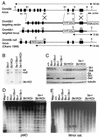
Dnmt3b6 has no enzymatic activity in vivo. (A) Strategy of targeted deletion of Dnmt3b exons 21 and 22. The top line shows the Dnmt3b genomic structure with exons represented by vertical bars. The targeting vector (second line) was constructed by replacing exons 21 and 22 with a PGK-puromycin cassette. A PGK-DTA cassette was introduced for negative selection to increase the targeting frequency. (B) Southern analysis of the genotype of ES cell lines. Genomic DNA was digested with _Eco_RV and hybridized to a 3′ external probe, as shown in (A). The 16-kb wild-type allele, the 5-kb Dnmt3b1 targeted allele, and the 14-kb Dnmt3b null allele (30) are indicated. (C) Lysates from the indicated cell lines were immunoblotted with anti-Dnmt3b (top), anti-Dnmt3a (middle), and anti-tubulin (bottom) antibodies. (D and E) Genomic DNA from the indicated ES cell lines was digested with _Hpa_II and hybridized to probes for endogenous C-type retrovirus repeats (D) and minor satellite repeats (E).
FIG. 6.
Active Dnmt3a/3b isoforms rescue the capacity of late-passage 7aabb cells to form terotomas in nude mice. (A) The indicated ES cell lines were injected into nude mice subcutaneously on both sides (three to four mice for each cell line, 5 × 105 cells per site), and the mice were examined for teratomas after 4 weeks. A typical representation of the size of the teratomas derived from each cell line is shown. (B) Histological sections of teratomas derived from J1, early-passage (P10) 7aabb, and Dnmt3a, Dnmt3a2, and Dnmt3b1 stable clones showing the presence of multiple types of differentiated cells.
FIG. 7.
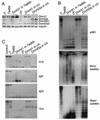
Dnmt1 and Dnmt3 proteins function cooperatively in maintaining methylation patterns. (A) Dnmt1 or Dnmt3a was overexpressed in 7aabb (P70) or Dnmt1−/− (c/c) ES cells as indicated and stable clones were examined for protein expression by immunoblotting with anti-Dnmt1 (top), anti-Dnmt3a (middle), and anti-tubulin (bottom) antibodies. (B and C) Genomic DNA from the indicated ES cell lines was analyzed for methylation of repetitive sequences (B) and unique genes (C) with the indicated probes.
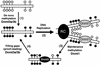
FIG. 8.
Model for the distinct roles of Dnmt1 and Dnmt3a/3b in de novo and maintenance methylation. (Step 1) Dnmt3a and Dnmt3b establish new DNA methylation patterns by de novo methylation of symmetric CpG dinucleotides (lollipops). (Step 2) Upon DNA replication, the new synthesized DNA becomes hemimethylated at CpG sites. (Step 3) Dnmt1, which is localized to the replication complex (RC), restores full methylation by methylating hemimethylated DNA. However, some CpG sites are left untouched by Dnmt1 and remain hemimethylated. (Step 4) Dnmt3a and Dnmt3b, which may also localize to the RC, recognize unmethylated CpG sites and restore methylation via de novo methylation. In summary, Dnmt1 is the major maintenance methyltransferase, and it has little or no de novo methylation activity in vivo. Dnmt3a and Dnmt3b possess active de novo methyltransferase activity in vivo and are essential for the establishment and maintenance of DNA methylation patterns.
Similar articles
- The inactive Dnmt3b3 isoform preferentially enhances Dnmt3b-mediated DNA methylation.
Zeng Y, Ren R, Kaur G, Hardikar S, Ying Z, Babcock L, Gupta E, Zhang X, Chen T, Cheng X. Zeng Y, et al. Genes Dev. 2020 Nov 1;34(21-22):1546-1558. doi: 10.1101/gad.341925.120. Epub 2020 Oct 1. Genes Dev. 2020. PMID: 33004415 Free PMC article. - Regulation of de novo and maintenance DNA methylation by DNA methyltransferases in postimplantation embryos.
Xu Z, Shi J, Chen Q, Yang S, Wang Z, Xiao B, Lai Z, Jing Y, Li Y, Li X. Xu Z, et al. J Biol Chem. 2025 Jan;301(1):107990. doi: 10.1016/j.jbc.2024.107990. Epub 2024 Nov 13. J Biol Chem. 2025. PMID: 39542247 Free PMC article. - Widespread recovery of methylation at gametic imprints in hypomethylated mouse stem cells following rescue with DNMT3A2.
Thakur A, Mackin SJ, Irwin RE, O'Neill KM, Pollin G, Walsh C. Thakur A, et al. Epigenetics Chromatin. 2016 Nov 22;9:53. doi: 10.1186/s13072-016-0104-2. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27895716 Free PMC article. - Tissue-specific roles of de novo DNA methyltransferases.
Tóth DM, Szeri F, Ashaber M, Muazu M, Székvölgyi L, Arányi T. Tóth DM, et al. Epigenetics Chromatin. 2025 Jan 17;18(1):5. doi: 10.1186/s13072-024-00566-2. Epigenetics Chromatin. 2025. PMID: 39819598 Free PMC article. Review. - Using human disease mutations to understand de novo DNA methyltransferase function.
Rolls W, Wilson MD, Sproul D. Rolls W, et al. Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review.
Cited by
- Dnmt3a is essential for hematopoietic stem cell differentiation.
Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, Liang S, Lu Y, Darlington GJ, Meissner A, Issa JP, Godley LA, Li W, Goodell MA. Challen GA, et al. Nat Genet. 2011 Dec 4;44(1):23-31. doi: 10.1038/ng.1009. Nat Genet. 2011. PMID: 22138693 Free PMC article. - Negative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancer.
Jia Y, Li P, Fang L, Zhu H, Xu L, Cheng H, Zhang J, Li F, Feng Y, Li Y, Li J, Wang R, Du JX, Li J, Chen T, Ji H, Han J, Yu W, Wu Q, Wong J. Jia Y, et al. Cell Discov. 2016 Apr 12;2:16007. doi: 10.1038/celldisc.2016.7. eCollection 2016. Cell Discov. 2016. PMID: 27462454 Free PMC article. - Lysine methyltransferase G9a is not required for DNMT3A/3B anchoring to methylated nucleosomes and maintenance of DNA methylation in somatic cells.
Sharma S, Gerke DS, Han HF, Jeong S, Stallcup MR, Jones PA, Liang G. Sharma S, et al. Epigenetics Chromatin. 2012 Jan 27;5(1):3. doi: 10.1186/1756-8935-5-3. Epigenetics Chromatin. 2012. PMID: 22284370 Free PMC article. - Genetic Studies on Mammalian DNA Methyltransferases.
Dan J, Chen T. Dan J, et al. Adv Exp Med Biol. 2022;1389:111-136. doi: 10.1007/978-3-031-11454-0_5. Adv Exp Med Biol. 2022. PMID: 36350508 Free PMC article. - Relationship between LTR methylation and gag expression of HIV-1 in human spermatozoa and sperm-derived embryos.
Li F, Li L, Zhong Y, Xie Q, Huang J, Kang X, Wang D, Xu L, Huang T. Li F, et al. PLoS One. 2013;8(1):e54801. doi: 10.1371/journal.pone.0054801. Epub 2013 Jan 28. PLoS One. 2013. PMID: 23382972 Free PMC article.
References
- Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. - PubMed
- Beaulieu, N., S. Morin, I. C. Chute, M.-F. Robert, H. Nguyen, and A. R. MacLeod. 2002. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J. Biol. Chem. 277:28176-28181. - PubMed
- Bestor, T., A. Laudano, R. Mattaliano, and V. Ingram. 1988. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells: the carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203:971-983. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases