A gene recommender algorithm to identify coexpressed genes in C. elegans - PubMed (original) (raw)
A gene recommender algorithm to identify coexpressed genes in C. elegans
Art B Owen et al. Genome Res. 2003 Aug.
Abstract
One of the most important uses of whole-genome expression data is for the discovery of new genes with similar function to a given list of genes (the query) already known to have closely related function. We have developed an algorithm, called the gene recommender, that ranks genes according to how strongly they correlate with a set of query genes in those experiments for which the query genes are most strongly coregulated. We used the gene recommender to find other genes coexpressed with several sets of query genes, including genes known to function in the retinoblastoma complex. Genetic experiments confirmed that one gene (JC8.6) identified by the gene recommender acts with lin-35 Rb to regulate vulval cell fates, and that another gene (wrm-1) acts antagonistically. We find that the gene recommender returns lists of genes with better precision, for fixed levels of recall, than lists generated using the C. elegans expression topomap.
Figures
Figure 1
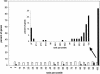
Some DNA microarray experiments show coregulation of the genes in the Rb query. Histogram of experiment scores obtained using the retinoblastoma query (black bars) or using 100 random queries of the same size (white bars). The arrow indicates the maximum experiment score obtained across all the randomizations. A substantial fraction of the experiment Z scores obtained using the retinoblastoma query were higher than the maximum Z score obtained from random queries.
Figure 2
Leave-one-out cross-validation. Histograms of percentile ranks obtained after removing a gene from a query list of genes, building a cassette around the remaining query genes, and then scoring the held-out gene. Open bars show the histogram obtained from random queries ranging in size from 4 to 50; black bars show the histogram obtained from all the queries used in this study. The inset is an expanded view of the highest-scoring genes.
Figure 3
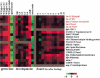
Expression profiles of genes in the Rb hit list. The germ line experiments compared expression in wild-type animals to glp-4 mutants lacking a germ line, and in mutants making only sperm to mutants making only oocytes (Reinke et al. 2000). The development experiments compared expression in whole wild-type worms throughout development and in hermaphrodites versus males (Jiang et al. 2001). The dauer experiments compared expression as animals exit the dauer stage following feeding in a timecourse experiment (Wang and Kim 2003). Rb query genes are shown in red. Scale shows level of expression.
Figure 4
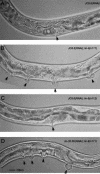
JC8.6(RNAi) results in a Muv phenotype similar to lin-35 Rb(RNAi). (A) JC8.6(RNAi), (B) JC8.6(RNAi) lin-8(n111), (C) JC8.6(RNAi) lin-9(n112), (D) lin-35 Rb(RNAi) lin-8(n111). Arrows point to the vulva in A and C, and to pseudovulvae in B and D. Adults were fed bacteria expressing JC8.6 dsRNA, and the phenotypes of their progeny were scored. Scale bar is 20 μM.
Similar articles
- A gene expression map for Caenorhabditis elegans.
Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. Kim SK, et al. Science. 2001 Sep 14;293(5537):2087-92. doi: 10.1126/science.1061603. Science. 2001. PMID: 11557892 - Comparative genomics of gene expression in the parasitic and free-living nematodes Strongyloides stercoralis and Caenorhabditis elegans.
Mitreva M, McCarter JP, Martin J, Dante M, Wylie T, Chiapelli B, Pape D, Clifton SW, Nutman TB, Waterston RH. Mitreva M, et al. Genome Res. 2004 Feb;14(2):209-20. doi: 10.1101/gr.1524804. Genome Res. 2004. PMID: 14762059 Free PMC article. - Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity.
Ceron J, Rual JF, Chandra A, Dupuy D, Vidal M, van den Heuvel S. Ceron J, et al. BMC Dev Biol. 2007 Apr 6;7:30. doi: 10.1186/1471-213X-7-30. BMC Dev Biol. 2007. PMID: 17417969 Free PMC article. - Functional exploration of the C. elegans genome using DNA microarrays.
Reinke V. Reinke V. Nat Genet. 2002 Dec;32 Suppl:541-6. doi: 10.1038/ng1039. Nat Genet. 2002. PMID: 12454651 Review. - Profiling C. elegans gene expression with DNA microarrays.
Portman DS. Portman DS. WormBook. 2006 Jan 20:1-11. doi: 10.1895/wormbook.1.104.1. WormBook. 2006. PMID: 18050445 Free PMC article. Review. No abstract available.
Cited by
- Query-based biclustering of gene expression data using Probabilistic Relational Models.
Zhao H, Cloots L, Van den Bulcke T, Wu Y, De Smet R, Storms V, Meysman P, Engelen K, Marchal K. Zhao H, et al. BMC Bioinformatics. 2011 Feb 15;12 Suppl 1(Suppl 1):S37. doi: 10.1186/1471-2105-12-S1-S37. BMC Bioinformatics. 2011. PMID: 21342568 Free PMC article. - Co-regulatory expression quantitative trait loci mapping: method and application to endometrial cancer.
Kompass KS, Witte JS. Kompass KS, et al. BMC Med Genomics. 2011 Jan 12;4:6. doi: 10.1186/1755-8794-4-6. BMC Med Genomics. 2011. PMID: 21226949 Free PMC article. - Integrative analysis of RUNX1 downstream pathways and target genes.
Michaud J, Simpson KM, Escher R, Buchet-Poyau K, Beissbarth T, Carmichael C, Ritchie ME, Schütz F, Cannon P, Liu M, Shen X, Ito Y, Raskind WH, Horwitz MS, Osato M, Turner DR, Speed TP, Kavallaris M, Smyth GK, Scott HS. Michaud J, et al. BMC Genomics. 2008 Jul 31;9:363. doi: 10.1186/1471-2164-9-363. BMC Genomics. 2008. PMID: 18671852 Free PMC article. - A molecular-properties-based approach to understanding PDZ domain proteins and PDZ ligands.
Giallourakis C, Cao Z, Green T, Wachtel H, Xie X, Lopez-Illasaca M, Daly M, Rioux J, Xavier R. Giallourakis C, et al. Genome Res. 2006 Aug;16(8):1056-72. doi: 10.1101/gr.5285206. Epub 2006 Jul 6. Genome Res. 2006. PMID: 16825666 Free PMC article. - Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex.
Harrison MM, Ceol CJ, Lu X, Horvitz HR. Harrison MM, et al. Proc Natl Acad Sci U S A. 2006 Nov 7;103(45):16782-7. doi: 10.1073/pnas.0608461103. Epub 2006 Oct 30. Proc Natl Acad Sci U S A. 2006. PMID: 17075059 Free PMC article.
References
- Brown, P.O. and Botstein, D. 1999. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 21: 33-37. - PubMed
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. - PubMed
- Ceol, C.J. and Horvitz, H.R. 2001. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol. Cell 7: 461-473. - PubMed
- Chase, D., Serafinas, C., Ashcroft, N., Kosinski, M., Longo, D., Ferris, D.K., and Golden, A. 2000. The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis 26: 26-41. - PubMed
WEB SITE REFERENCES
- http://pmgm2.stanford.edu/~kimlab/cassettes; details of the gene recommender and a Web interface to our software and data.
- http://www-stat.stanford.edu/~owen/transposable; articles and links comparing data analysis of DNA expression, recommender engines, search engines, and educational testing.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources