Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes - PubMed (original) (raw)
Comparative Study
. 2003 Aug;13(8):1863-72.
doi: 10.1101/gr.1272403.
Affiliations
- PMID: 12902380
- PMCID: PMC403777
- DOI: 10.1101/gr.1272403
Comparative Study
Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes
Edward Yang et al. Genome Res. 2003 Aug.
Abstract
Although mRNA decay rates are a key determinant of the steady-state concentration for any given mRNA species, relatively little is known, on a population level, about what factors influence turnover rates and how these rates are integrated into cellular decisions. We decided to measure mRNA decay rates in two human cell lines with high-density oligonucleotide arrays that enable the measurement of decay rates simultaneously for thousands of mRNA species. Using existing annotation and the Gene Ontology hierarchy of biological processes, we assign mRNAs to functional classes at various levels of resolution and compare the decay rate statistics between these classes. The results show statistically significant organizational principles in the variation of decay rates among functional classes. In particular, transcription factor mRNAs have increased average decay rates compared with other transcripts and are enriched in "fast-decaying" mRNAs with half-lives <2 h. In contrast, we find that mRNAs for biosynthetic proteins have decreased average decay rates and are deficient in fast-decaying mRNAs. Our analysis of data from a previously published study of Saccharomyces cerevisiae mRNA decay shows the same functional organization of decay rates, implying that it is a general organizational scheme for eukaryotes. Additionally, we investigated the dependence of decay rates on sequence composition, that is, the presence or absence of short mRNA motifs in various regions of the mRNA transcript. Our analysis recovers the positive correlation of mRNA decay with known AU-rich mRNA motifs, but we also uncover further short mRNA motifs that show statistically significant correlation with decay. However, we also note that none of these motifs are strong predictors of mRNA decay rate, indicating that the regulation of mRNA decay is more complex and may involve the cooperative binding of several RNA-binding proteins at different sites.
Figures
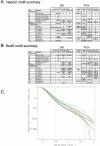
Figure 1
Functional analysis of decaying transcripts in human cells. (A,B) Probe sets from the HepG2 experiments (A) or the Bud8 experiment (B) were grouped by functional (i.e., Gene Ontology, GO) category, and both decay rate and the percentage of fast decayers were inferred using procedures we call DRI and PFDI (see Methods: Statistical Analysis and Decay Rate Calculations). For DRI, the average decay rates were calculated (error bars denote 99% posterior probability interval (PPI)) for probe sets corresponding to the functional category listed in the center (Group, blue). If the GO category in question was separated from the rest of the probe sets (Nongroup, purple) with >99% probability (see _p_-values to left), the distribution was described as “FASTER” or “SLOWER” as appropriate. Otherwise, the GO distribution was said to be “NO SIG” (not significantly) different from the other probe sets. For PFDI, the percentage of probe sets (error bars again denote 99% PPI) decaying with a rate >0.5 h-1 (2 h half-life) were calculated for probe sets inside of the stated GO category (Group) or outside the category (Nongroup). If the GO category's probe sets were enriched/depleted in the rapid turnover pool with at least 99% probability (see _p_-values to the right), the category was said to be “OVER” (overrepresented) or “UNDER” (underrepresented), respectively. Otherwise, the category was listed as “NO SIG” (no significant) enrichment. For comparison, the same analysis (“MANUAL”) was performed using a set of probe sets corresponding to SWISS-PROT entries annotated as transcription-related (see Methods). (C) Reverse cumulative distribution of decay rates for probe sets in different functional classes (HepG2 experiments). Decay rate r is shown horizontally, while vertically the fraction of probe sets with decay rates higher than r is plotted on a logarithmicscale. The pairs of lines show the 98% posterior probability intervals for the fraction at each value of r. (Red) GO process transcription; (black) all probe sets; (green) biosynthesis. The gray line indicates the decay rate r = 0.5 h-1, which is our cutoff for fast decay in PFDI.
Figure 1
Functional analysis of decaying transcripts in human cells. (A,B) Probe sets from the HepG2 experiments (A) or the Bud8 experiment (B) were grouped by functional (i.e., Gene Ontology, GO) category, and both decay rate and the percentage of fast decayers were inferred using procedures we call DRI and PFDI (see Methods: Statistical Analysis and Decay Rate Calculations). For DRI, the average decay rates were calculated (error bars denote 99% posterior probability interval (PPI)) for probe sets corresponding to the functional category listed in the center (Group, blue). If the GO category in question was separated from the rest of the probe sets (Nongroup, purple) with >99% probability (see _p_-values to left), the distribution was described as “FASTER” or “SLOWER” as appropriate. Otherwise, the GO distribution was said to be “NO SIG” (not significantly) different from the other probe sets. For PFDI, the percentage of probe sets (error bars again denote 99% PPI) decaying with a rate >0.5 h-1 (2 h half-life) were calculated for probe sets inside of the stated GO category (Group) or outside the category (Nongroup). If the GO category's probe sets were enriched/depleted in the rapid turnover pool with at least 99% probability (see _p_-values to the right), the category was said to be “OVER” (overrepresented) or “UNDER” (underrepresented), respectively. Otherwise, the category was listed as “NO SIG” (no significant) enrichment. For comparison, the same analysis (“MANUAL”) was performed using a set of probe sets corresponding to SWISS-PROT entries annotated as transcription-related (see Methods). (C) Reverse cumulative distribution of decay rates for probe sets in different functional classes (HepG2 experiments). Decay rate r is shown horizontally, while vertically the fraction of probe sets with decay rates higher than r is plotted on a logarithmicscale. The pairs of lines show the 98% posterior probability intervals for the fraction at each value of r. (Red) GO process transcription; (black) all probe sets; (green) biosynthesis. The gray line indicates the decay rate r = 0.5 h-1, which is our cutoff for fast decay in PFDI.
Figure 1
Functional analysis of decaying transcripts in human cells. (A,B) Probe sets from the HepG2 experiments (A) or the Bud8 experiment (B) were grouped by functional (i.e., Gene Ontology, GO) category, and both decay rate and the percentage of fast decayers were inferred using procedures we call DRI and PFDI (see Methods: Statistical Analysis and Decay Rate Calculations). For DRI, the average decay rates were calculated (error bars denote 99% posterior probability interval (PPI)) for probe sets corresponding to the functional category listed in the center (Group, blue). If the GO category in question was separated from the rest of the probe sets (Nongroup, purple) with >99% probability (see _p_-values to left), the distribution was described as “FASTER” or “SLOWER” as appropriate. Otherwise, the GO distribution was said to be “NO SIG” (not significantly) different from the other probe sets. For PFDI, the percentage of probe sets (error bars again denote 99% PPI) decaying with a rate >0.5 h-1 (2 h half-life) were calculated for probe sets inside of the stated GO category (Group) or outside the category (Nongroup). If the GO category's probe sets were enriched/depleted in the rapid turnover pool with at least 99% probability (see _p_-values to the right), the category was said to be “OVER” (overrepresented) or “UNDER” (underrepresented), respectively. Otherwise, the category was listed as “NO SIG” (no significant) enrichment. For comparison, the same analysis (“MANUAL”) was performed using a set of probe sets corresponding to SWISS-PROT entries annotated as transcription-related (see Methods). (C) Reverse cumulative distribution of decay rates for probe sets in different functional classes (HepG2 experiments). Decay rate r is shown horizontally, while vertically the fraction of probe sets with decay rates higher than r is plotted on a logarithmicscale. The pairs of lines show the 98% posterior probability intervals for the fraction at each value of r. (Red) GO process transcription; (black) all probe sets; (green) biosynthesis. The gray line indicates the decay rate r = 0.5 h-1, which is our cutoff for fast decay in PFDI.
Figure 2
Motif analysis of decaying transcripts in human cells. (A,B) The probe sets from the four HepG2 experiments (A) or the Bud8 experiment (B) were analyzed for the relationship between transcript decay and the presence of particular sequence motifs. The results of DRI and PFDI (same procedures used in Fig. 1) are summarized in A and B. For the motif analysis, we performed separate inferences for portions of the sequence (3′-UTR, 5′-UTR, ORF) and the cDNA sequence considered as a whole. For DRI, we compared the average decay rate of the probe sets from genes containing the motif in a specified location with rates of all other probe sets: significant (99% probability or greater) increases are shown in bold and significant decreases in italics. For PFDI, motifs that are overrepresented in the rapidly decaying transcript pool (r > 0.5 h-1) when located in a given position are shown in bold; underrepresented transcripts are shown in italics (again, 99% probability cutoff for both). Motif–location combinations without statistically significant changes are shown as blank, and combinations with too few probe pairs for inference (≤25 probe pairs) are indicated with “n.a.” For more details on the motif analysis (e.g., extent of shift in average decay rate, percent enrichment), see Supplemental Tables 4–6. (C) Reverse cumulative distribution of decay rates for probe sets from genes that contain particular sequence motifs in their 3′-UTR (HepG2 experiment). Decay rate r is shown horizontally, while vertically the fraction of probe sets with decay rates higher than r is plotted on a logarithmicscale. The pairs of lines show the 98% posterior probability intervals for the fraction at each value of r. (Red) Motif 1; (blue) motif MEGSHORT; (green) Motif 2E; (light green) Motif H1; (black) all probe sets. “Described” AU-rich decay motifs (1–2E, MEG, MEGSHORT) and “undescribed” motifs were derived from the sources mentioned in Methods.
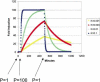
Figure 3
Simulation of the effect of decay rate on gene induction. Equation 6 (Methods) was solved for a step function RNA synthesis impulse: the synthesis rate was increased from 1 copy/(min · cell) to 100 copies/(min · cell) from 50–550 min. Using the indicated first-order decay constant (in units of 1/minute), the steady-state concentration of RNA was calculated and divided by initial concentration to determine the fold induction.
Similar articles
- Quantitation of alpha-fetoprotein and albumin messenger RNA in human hepatocellular carcinoma.
Niwa Y, Matsumura M, Shiratori Y, Imamura M, Kato N, Shiina S, Okudaira T, Ikeda Y, Inoue T, Omata M. Niwa Y, et al. Hepatology. 1996 Jun;23(6):1384-92. doi: 10.1053/jhep.1996.v23.pm0008675155. Hepatology. 1996. PMID: 8675155 - Global analysis of mRNA decay in Halobacterium salinarum NRC-1 at single-gene resolution using DNA microarrays.
Hundt S, Zaigler A, Lange C, Soppa J, Klug G. Hundt S, et al. J Bacteriol. 2007 Oct;189(19):6936-44. doi: 10.1128/JB.00559-07. Epub 2007 Jul 20. J Bacteriol. 2007. PMID: 17644597 Free PMC article. - Mybl2 expression is under genetic control and contributes to determine a hepatocellular carcinoma susceptible phenotype.
Frau M, Ladu S, Calvisi DF, Simile MM, Bonelli P, Daino L, Tomasi ML, Seddaiu MA, Feo F, Pascale RM. Frau M, et al. J Hepatol. 2011 Jul;55(1):111-9. doi: 10.1016/j.jhep.2010.10.031. Epub 2010 Dec 7. J Hepatol. 2011. PMID: 21419759 - Inhibiting transcription in cultured metazoan cells with actinomycin D to monitor mRNA turnover.
Lai WS, Arvola RM, Goldstrohm AC, Blackshear PJ. Lai WS, et al. Methods. 2019 Feb 15;155:77-87. doi: 10.1016/j.ymeth.2019.01.003. Epub 2019 Jan 6. Methods. 2019. PMID: 30625384 Free PMC article. Review. - mRNA stability in mammalian cells.
Ross J. Ross J. Microbiol Rev. 1995 Sep;59(3):423-50. doi: 10.1128/mr.59.3.423-450.1995. Microbiol Rev. 1995. PMID: 7565413 Free PMC article. Review.
Cited by
- Modeling RNA degradation for RNA-Seq with applications.
Wan L, Yan X, Chen T, Sun F. Wan L, et al. Biostatistics. 2012 Sep;13(4):734-47. doi: 10.1093/biostatistics/kxs001. Epub 2012 Feb 21. Biostatistics. 2012. PMID: 22353193 Free PMC article. - Metabolic states following accumulation of intracellular aggregates: implications for neurodegenerative diseases.
Vazquez A. Vazquez A. PLoS One. 2013 May 7;8(5):e63822. doi: 10.1371/journal.pone.0063822. Print 2013. PLoS One. 2013. PMID: 23667676 Free PMC article. - Cell uptake and intracellular trafficking of bioreducible poly(amidoamine) nanoparticles for efficient mRNA translation in chondrocytes.
Pontes AP, van der Wal S, Ranamalla SR, Roelofs K, Tomuta I, Creemers LB, Rip J. Pontes AP, et al. Front Bioeng Biotechnol. 2023 Nov 10;11:1290871. doi: 10.3389/fbioe.2023.1290871. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 38026902 Free PMC article. - Retention of exogenous mRNAs selectively in the germ cells of the sea urchin requires only a 5'-cap and a 3'-UTR.
Oulhen N, Wessel GM. Oulhen N, et al. Mol Reprod Dev. 2013 Jul;80(7):561-9. doi: 10.1002/mrd.22193. Epub 2013 Jun 27. Mol Reprod Dev. 2013. PMID: 23686945 Free PMC article. - Circular RNA Expression and Interaction Patterns Are Perturbed in Amyotrophic Lateral Sclerosis.
Aquilina-Reid C, Brennan S, Curry-Hyde A, Teunisse GM, The Nygc Als Consortium, Janitz M. Aquilina-Reid C, et al. Int J Mol Sci. 2022 Nov 24;23(23):14665. doi: 10.3390/ijms232314665. Int J Mol Sci. 2022. PMID: 36498994 Free PMC article.
References
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451-464. - PubMed
- Darnell Jr., J.E. 1982. Variety in the level of gene control in eukaryotic cells. Nature 297: 365-371. - PubMed
WEB SITE REFERENCES
- http://genomes.rockefeller.edu/~yange/; decay rate estimates for 5245 accessions.
- http://genome-www.stanford.edu/Saccharomyces; Saccharomyces cerevisiae database.
- http://genome-www.stanford.edu/turnover; turnover cDNA microarray data from Saccharomyces cerevisiae.
- http://www.affymetrix.com/analysis/index.affx; Affymetrix.
- http://www.geneontology.org; Gene Ontology Consortium.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources