Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis - PubMed (original) (raw)
Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis
Scott D Michaels et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Plant species have evolved a wide variety of flowering habits, each adapted to maximize reproductive success in their local environment. Even within a species, accessions from different environments can exhibit markedly different flowering behavior. In Arabidopsis, some accessions are rapid-cycling summer annuals, whereas others accessions are late flowering and vernalization responsive and thus behave as winter annuals. Two genes, FLOWERING LOCUS C (FLC) and FRIGIDA (FRI), interact synergistically to confer the winter-annual habit. Previous work has shown that many summer-annual accessions contain null mutations in the FRI gene; thus it appears that these summer-annual accessions have arisen from winter-annual ancestors by losing FRI function. In this work we demonstrate that naturally occurring allelic variation in FLC has provided another route to the evolution of summer-annual flowering behavior in Arabidopsis. We have identified two summer-annual accessions, Da (1)-12 and Shakhdara, that contain functional alleles of FRI, but are early flowering because of weak alleles of FLC. We have also determined that the weak allele of FLC found in Landsberg erecta is naturally occurring. Unlike accessions that have arisen because of loss-of-function mutations in FRI, the FLC alleles from Da (1)-12, Shakhdara, and Landsberg erecta are not nulls; however, they exhibit lower steady-state mRNA levels than strong alleles of FLC. Sequence analysis indicates that these weak alleles of FLC have arisen independently at least twice during the course of evolution.
Figures
Fig. 1.
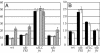
Determination of FRI and FLC activity in early-flowering accessions. (A) Da (1)-12 (open bars), Shakhdara (gray bars), and Kondara (cross-hatched bars) were crossed to tester lines homozygous for the indicated genotypes, and the rosette leaf numbers of the resulting F1 plants are presented. A line homozygous for FRI and_flc_-3 was used as a control (black bars). (B)L_er_ (black bars) and LER (open bars) were crossed to tester lines homozygous for the indicated genotypes, and the rosette leaf numbers of the resulting F1 plants are presented.
Fig. 2.
Transformation with a genomic FLC construct is sufficient to transform Da (1)-12 and Shakhdara from summer annuals to winter annuals.
Fig. 3.
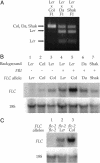
Analysis of FLC mRNA levels. (A) Allele-specific RT-PCR was used to examine the relative expression levels of different FLC alleles in F1 plants resulting from the indicated crosses. RT-PCR products were digested with Hpy188III, resolved on a 1.5% agarose gel, and visualized by staining with ethidium bromide. A unique Hpy188III site in the L_er_ RT-PCR product allows the L_er_ product to be separated from the Col, Da (1)-12, and Shakhdara products. (B) RNA blot analysis of FLC expression in L_er_, Col, Da (1)-12, and Shakhdara, and in near isogenic lines containing FRI,FLC_-Col, or FRI and FLC_-Col in the L_er genetic background. (C) RNA blot analysis of FLC expression in F1 plants resulting from crosses between a line containing_FRI and flc_-2 in the Col background and L_er or a near isogenic line containing FLC_-Col in the L_er genetic background. The _flc_-2 allele of FLC is a large deletion that completely removes the FLC gene.
Fig. 4.
Summary of the polymorphisms detected in the first intron of FLC in L_er_ and Da (1)-12. The structure of the Col allele of FLC is shown with exons as black boxes and introns as open boxes. L_er_ and Da (1)-12 insertions and the L_er_ 30-bp deletion are shown as gray boxes.
Fig. 5.
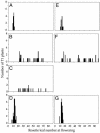
Effect of L_er_ polymorphisms on FLC activity in a_FRI_-containing or autonomous-pathway-mutant background. The number of rosette leaves formed by the primary meristem was determined for a line homozygous for FRI and flc_-3 in the Col background alone (A) or after transformation with a wild-type Col FLC genomic construct (B), a Col FLC genomic construct containing the 30-bp deletion found in L_er (C), or a Col FLC genomic construct containing the 1.2-kb insertion found in L_er_ (D). In addition, rosette leaves formed before flowering were determined for a line homozygous for _fpa_-7 and flc_-3 either alone (E) or after transformation with a wild-type Col FLC genomic construct (F) or a Col FLC genomic construct containing the 1.2-kb insertion found in L_er (G).
Similar articles
- Analysis of the molecular basis of flowering time variation in Arabidopsis accessions.
Gazzani S, Gendall AR, Lister C, Dean C. Gazzani S, et al. Plant Physiol. 2003 Jun;132(2):1107-14. doi: 10.1104/pp.103.021212. Epub 2003 May 22. Plant Physiol. 2003. PMID: 12805638 Free PMC article. - Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis.
Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C. Shindo C, et al. Plant Physiol. 2005 Jun;138(2):1163-73. doi: 10.1104/pp.105.061309. Epub 2005 May 20. Plant Physiol. 2005. PMID: 15908596 Free PMC article. - FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions.
Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D. Werner JD, et al. Genetics. 2005 Jul;170(3):1197-207. doi: 10.1534/genetics.104.036533. Epub 2005 May 23. Genetics. 2005. PMID: 15911588 Free PMC article. - Vernalization: a model for investigating epigenetics and eukaryotic gene regulation in plants.
Schmitz RJ, Amasino RM. Schmitz RJ, et al. Biochim Biophys Acta. 2007 May-Jun;1769(5-6):269-75. doi: 10.1016/j.bbaexp.2007.02.003. Epub 2007 Feb 27. Biochim Biophys Acta. 2007. PMID: 17383745 Review. - Role of chromatin modification in flowering-time control.
He Y, Amasino RM. He Y, et al. Trends Plant Sci. 2005 Jan;10(1):30-5. doi: 10.1016/j.tplants.2004.11.003. Trends Plant Sci. 2005. PMID: 15642521 Review.
Cited by
- Analysis of ambient temperature-responsive transcriptome in shoot apical meristem of heat-tolerant and heat-sensitive broccoli inbred lines during floral head formation.
Lin CW, Fu SF, Liu YJ, Chen CC, Chang CH, Yang YW, Huang HJ. Lin CW, et al. BMC Plant Biol. 2019 Jan 3;19(1):3. doi: 10.1186/s12870-018-1613-x. BMC Plant Biol. 2019. PMID: 30606114 Free PMC article. - Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles.
Olsen KM, Halldorsdottir SS, Stinchcombe JR, Weinig C, Schmitt J, Purugganan MD. Olsen KM, et al. Genetics. 2004 Jul;167(3):1361-9. doi: 10.1534/genetics.103.024950. Genetics. 2004. PMID: 15280248 Free PMC article. - Environmental and molecular analysis of the floral transition in the lower eudicot Aquilegia formosa.
Ballerini ES, Kramer EM. Ballerini ES, et al. Evodevo. 2011 Feb 17;2:4. doi: 10.1186/2041-9139-2-4. Evodevo. 2011. PMID: 21329499 Free PMC article. - A Molecular switch for FLOWERING LOCUS C activation determines flowering time in Arabidopsis.
Shen L, Zhang Y, Sawettalake N. Shen L, et al. Plant Cell. 2022 Feb 3;34(2):818-833. doi: 10.1093/plcell/koab286. Plant Cell. 2022. PMID: 34850922 Free PMC article. - Fine-scale mapping of natural variation in fly fecundity identifies neuronal domain of expression and function of an aquaporin.
Bergland AO, Chae HS, Kim YJ, Tatar M. Bergland AO, et al. PLoS Genet. 2012;8(4):e1002631. doi: 10.1371/journal.pgen.1002631. Epub 2012 Apr 5. PLoS Genet. 2012. PMID: 22509142 Free PMC article.
References
- Cumming, B. G. (1969) Can. J. Bot. 41, 901–926.
- Michaels, S. & Amasino, R. (2000) Plant Cell Environ. 23, 1145–1154.
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R. & Dean, C. (2000) Science 290, 344–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases