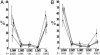High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes - PubMed (original) (raw)
High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes
Elena Allen et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Genes subject to monoallelic expression are expressed from only one of the two alleles either selected at random (random monoallelic genes) or in a parent-of-origin specific manner (imprinted genes). Because high densities of long interspersed nuclear element (LINE)-1 transposon sequence have been implicated in X-inactivation, we asked whether monoallelically expressed autosomal genes are also flanked by high densities of LINE-1 sequence. A statistical analysis of repeat content in the regions surrounding monoallelically and biallelically expressed genes revealed that random monoallelic genes were flanked by significantly higher densities of LINE-1 sequence, evolutionarily more recent and less truncated LINE-1 elements, fewer CpG islands, and fewer base-pairs of short interspersed nuclear elements (SINEs) sequence than biallelically expressed genes. Random monoallelic and imprinted genes were pooled and subjected to a clustering analysis algorithm, which found two clusters on the basis of aforementioned sequence characteristics. Interestingly, these clusters did not follow the random monoallelic vs. imprinted classifications. We infer that chromosomal sequence context plays a role in monoallelic gene expression and may involve the recognition of long repeats or other features. The sequence characteristics that distinguished the high-LINE-1 category were used to identify more than 1,000 additional genes from the human and mouse genomes as candidate genes for monoallelic expression.
Figures
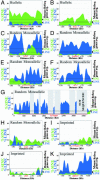
Fig. 1.
LINE-1 and SINE sequence abundance in regions flanking biallelically expressed, random monoallelic, and imprinted genes. Percent LINE-1 and SINE were calculated as the quotient of base pairs of repeats to total base pairs as described (see supporting information). Graphs for percent LINE-1 and SINE sequence are superimposed, with the higher value behind the lower value. Green, SINE sequence content; blue, LINE-1 sequence content; red box, gene; stippled red line, Ig gene region; gray box, region for which sequence is not available. The overall LINE-1 and SINE averages for 200-kb regions flanking all available mouse or human RefSeq genes (see Methods) are marked to the right of each plot. LINE-1 average, blue letters; SINE average, green letters. Biallelically expressed genes: human β_-ACTIN_ (A) and mouse Rras-1 (B). Random monoallelically expressed genes: human XIST (C); mouse Ly49G2 (D); mouse olfactory gene cluster containing Or8,Or10, and Or28 (E); human IL-2 (F); mouse Ig κ gene region (G); mouse Jsap (H). Imprinted genes: mouse p57 (kip2) (I); human_H19_ (J); mouse Zfp127 (K).
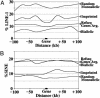
Fig. 2.
Average LINE-1 and SINE sequence abundance in regions flanking genes that are or are not subject to monoallelic expression. Percent LINE-1 or SINE sequence in sequential 20-kb sequence windows proceeding 100 kb upstream of the 5′ end of the gene, or extending 100 kb downstream from the 3′ end of the gene, are shown. Densities were calculated as described in_Methods_. (A) LINE-1 sequence averages. (B) SINE sequence averages.
Fig. 3.
Boxplots of select covariates. B, biallelically expressed genes; I, imprinted genes; RM, random monoallelically expressed genes; S, random sampling of 150 genes. The width of each box plot reflects the sample size.P values are determined by the Kruskal–Wallis test comparing B, I, and RM genes.
Fig. 4.
Distribution of LINE-1 ages between groups of genes. Plotted are the L1 subfamily mean percentages and their 95% confidence intervals. Below each LINE-1 type are P values across the corresponding values for the biallelic (white circle), imprinted (triangle), and random monoallelic (black circle) genes (Kruskal–Wallis test). Average (see Methods) for a random sample of 150 genes (squares). Mean percentages were calculated as the quotient of subfamily elements and total L1 elements. (A) Five mouse subfamilies are listed from the most ancient (L1M4) to the most recent (Lx). (B) Five human L1 subfamilies listed left to right from the evolutionarily most ancient (L1M4) repeats to the most recent (L1P). See supporting information for more details.
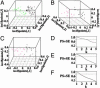
Fig. 5.
Isotonic multidimensional scaling plots (A–C) and prediction strength plots (D–F) of the 39 imprinted and 33 randomly inactivated genes in Table 1. (A) Mouse (green) and human (black) genes. (B) Mouse genes: black, cluster 1; red, cluster 2. (C) Human genes: black, cluster 1; red, cluster 2. We estimated the number of gene clusters using the prediction strength criterion: the number of clusters is estimated as the largest_k_ such that prediction strength (PS)+SE is >0.8 (dashed line). Prediction strengths plus their standard errors for different choices of the number of clusters (k) are shown. (D) Mouse and human genes. (E) Mouse genes. (F) Human genes.
Similar articles
- Short interspersed element (SINE) depletion and long interspersed element (LINE) abundance are not features universally required for imprinting.
Cowley M, de Burca A, McCole RB, Chahal M, Saadat G, Oakey RJ, Schulz R. Cowley M, et al. PLoS One. 2011 Apr 20;6(4):e18953. doi: 10.1371/journal.pone.0018953. PLoS One. 2011. PMID: 21533089 Free PMC article. - The distribution and conservation of retrotransposable elements in cattle.
Khatib H, Kim ES. Khatib H, et al. Epigenetics. 2008 Mar-Apr;3(2):81-8. doi: 10.4161/epi.3.2.5833. Epub 2008 Mar 3. Epigenetics. 2008. PMID: 18536529 - Tandem repeats in the CpG islands of imprinted genes.
Hutter B, Helms V, Paulsen M. Hutter B, et al. Genomics. 2006 Sep;88(3):323-32. doi: 10.1016/j.ygeno.2006.03.019. Epub 2006 May 11. Genomics. 2006. PMID: 16690248 - Epigenetic events regulating monoallelic gene expression.
Massah S, Beischlag TV, Prefontaine GG. Massah S, et al. Crit Rev Biochem Mol Biol. 2015;50(4):337-58. doi: 10.3109/10409238.2015.1064350. Epub 2015 Jul 9. Crit Rev Biochem Mol Biol. 2015. PMID: 26155735 Review. - The influence of DNA methylation on monoallelic expression.
da Rocha ST, Gendrel AV. da Rocha ST, et al. Essays Biochem. 2019 Dec 20;63(6):663-676. doi: 10.1042/EBC20190034. Essays Biochem. 2019. PMID: 31782494 Free PMC article. Review.
Cited by
- Assembly and analysis of the mouse immunoglobulin kappa gene sequence.
Brekke KM, Garrard WT. Brekke KM, et al. Immunogenetics. 2004 Oct;56(7):490-505. doi: 10.1007/s00251-004-0659-0. Epub 2004 Sep 18. Immunogenetics. 2004. PMID: 15378297 - Characterization of bovine (Bos taurus) imprinted genes from genomic to amino acid attributes by data mining approaches.
Karami K, Zerehdaran S, Javadmanesh A, Shariati MM, Fallahi H. Karami K, et al. PLoS One. 2019 Jun 6;14(6):e0217813. doi: 10.1371/journal.pone.0217813. eCollection 2019. PLoS One. 2019. PMID: 31170205 Free PMC article. - Differential Allelic Expression among Long Non-Coding RNAs.
Heskett MB, Spellman PT, Thayer MJ. Heskett MB, et al. Noncoding RNA. 2021 Oct 22;7(4):66. doi: 10.3390/ncrna7040066. Noncoding RNA. 2021. PMID: 34698262 Free PMC article. Review. - Short interspersed element (SINE) depletion and long interspersed element (LINE) abundance are not features universally required for imprinting.
Cowley M, de Burca A, McCole RB, Chahal M, Saadat G, Oakey RJ, Schulz R. Cowley M, et al. PLoS One. 2011 Apr 20;6(4):e18953. doi: 10.1371/journal.pone.0018953. PLoS One. 2011. PMID: 21533089 Free PMC article. - Epigenetic control of chromosome-associated lncRNA genes essential for replication and stability.
Heskett MB, Vouzas AE, Smith LG, Yates PA, Boniface C, Bouhassira EE, Spellman PT, Gilbert DM, Thayer MJ. Heskett MB, et al. Nat Commun. 2022 Oct 22;13(1):6301. doi: 10.1038/s41467-022-34099-7. Nat Commun. 2022. PMID: 36273230 Free PMC article.
References
- Nicholls, R. D. & Knepper, J. L. (2001) Annu. Rev. Genom. Hum. Genet. 2, 153–175. - PubMed
- Lyon, M. F. (1961) Nature 190, 372–373. - PubMed
- Riggs, A. D. (1990) Aust. J. Zool. 37, 419–441.
- Gartler, S. M. & Riggs, A. D. (1983) Annu. Rev. Genet. 17, 155–190. - PubMed
- Lyon, M. F. (1998) Cytogenet. Cell Genet. 80, 133–137. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases