Mpi recombinase globally modulates the surface architecture of a human commensal bacterium - PubMed (original) (raw)
Mpi recombinase globally modulates the surface architecture of a human commensal bacterium
Michael J Coyne et al. Proc Natl Acad Sci U S A. 2003.
Abstract
The mammalian gut represents a complex and diverse ecosystem, consisting of unique interactions between the host and microbial residents. Bacterial surfaces serve as an interface that promotes and responds to this dynamic exchange, a process essential to the biology of both symbionts. The human intestinal microorganism, Bacteroides fragilis, is able to extensively modulate its surface. Analysis of the B. fragilis genomic sequence, together with genetic conservation analyses, cross-species cloning experiments, and mutational studies, revealed that this organism utilizes an endogenous DNA inversion factor to globally modulate the expression of its surface structures. This DNA invertase is necessary for the inversion of at least 13 regions located throughout the genome, including the promoter regions for seven of the capsular polysaccharide biosynthesis loci, an accessory polysaccharide biosynthesis locus, and five other regions containing consensus promoter sequences. Bacterial DNA invertases of the serine site-specific recombinase family are typically encoded by imported elements such as phage and plasmids, and act locally on a single region of the imported element. In contrast, the conservation and unique global regulatory nature of the process in B. fragilis suggest an evolutionarily ancient mechanism for surface adaptation to the changing intestinal milieu during commensalism.
Figures
Fig. 1.
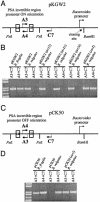
Inversion of the PSA promoter region by Ssr2. (A) Plasmid pKGW2 was constructed to monitor inversion of PSA promoter region from ON to OFF. The PSA promoter was cloned into the _Pst_I site of pFD340, and its inversion to the OFF orientation was monitored by PCR using primers A4 and C7. Site-specific recombinases were cloned into the _Bam_HI site, where their transcription is governed by a constitutive plasmid-borne promoter. (B) Ethidium bromide (EtBr)-stained agarose gel demonstrating the ability or inability of various candidate site-specific recombinases to invert the PSA promoter region. (C) Plasmid pCK50 was used to monitor inversion of the PSA promoter from the OFF orientation to the ON orientation. (D) EtBr-stained agarose gel demonstrating the ability of Ssr2 to invert the PSA promoter from the OFF orientation to the ON orientation.
Fig. 3.
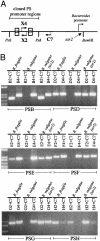
Direct role of Ssr2 (Mpi) in inversion of the PSB, PSD, PSE, PSF, PSG, and PSH promoter regions. (A) Diagrammatic representation of the PCR protocol that was used to detect inversion of each of six polysaccharide promoter regions when cloned into pFD340 and transferred to B. fragilis or B. vulgatus, with or without ssr2. The sequences of the primers used for each PCR are listed in Table 2. (B) EtBr-stained agarose gels demonstrating that each of the six promoter regions is able to invert in B. vulgatus only when ssr2 is present.
Fig. 2.
Ssr2 is necessary for inversion of all seven polysaccharide promoter regions. (A) Diagrammatic representation of the PCR method used to detect DNA inversions of each of the seven invertible promoter regions. Each of the primers contained within the IRs was used in a PCR with both the upstream and downstream primers individually. (B) EtBr-stained agarose gel demonstrating that the promoters of each of the seven polysaccharide biosynthesis loci is present in only a single orientation in the Δ_ssr2_ mutants relative to wild type. Three distinct locked polysaccharide promoter patterns were detected in the panel of eight mutants exemplified by Δ_ssr2_ mutants 2, 8, and 44. The bottom panel shows complementation of the locked genotypes when ssr2 is added in trans to Δ_ssr2_ mutant 44 (pKGW4R). (C) Western blots demonstrating the polysaccharide phenotypes of the three representative Δ_ssr2_ mutants.
Fig. 4.
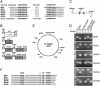
Characterization of six additional loci whose DNA inversions are controlled by Mpi. (A) The top line represents the upstream and downstream core consensus Mpi recognition sequence (ARACGTTCGTN{90,250}ACGAACGTYT) used to query the B. fragilis genome database. The IR sequences of the six new Mpi-controlled regions are shown. The bottom line shows the revised Mpi consensus recognition sequence based on these new additions. (B) Diagrammatic representation of the four new genomic regions containing six invertible regions and the immediate downstream ORFs. Genes highlighted in similar shades of gray are homologous to each other. IRs are shown as small boxes. (C) Design of the PCR used to examine each of the MCRs for inversion. Small boxes represent the upstream and downstream IR of each of the six regions. (D) EtBr-stained agarose gel demonstrating that each MCR undergoes inversion regulated by Mpi. (E) A scaled representation of the 5,205,140-bp B. fragilis 9343 genome demonstrating the relative locations of mpi and each of the Mpi-controlled regions. (F) Consensus promoter sequence present in each of the MCR invertible regions. The top line shows the B. fragilis consensus promoter sequence. The last nucleotide reported for each regions is adjacent to the first nucleotide of the downstream IR.
Similar articles
- Bacteroides fragilis synthesizes a DNA invertase affecting both a local and a distant region.
Roche-Hakansson H, Chatzidaki-Livanis M, Coyne MJ, Comstock LE. Roche-Hakansson H, et al. J Bacteriol. 2007 Mar;189(5):2119-24. doi: 10.1128/JB.01362-06. Epub 2006 Dec 22. J Bacteriol. 2007. PMID: 17189372 Free PMC article. - Tyrosine site-specific recombinases mediate DNA inversions affecting the expression of outer surface proteins of Bacteroides fragilis.
Weinacht KG, Roche H, Krinos CM, Coyne MJ, Parkhill J, Comstock LE. Weinacht KG, et al. Mol Microbiol. 2004 Sep;53(5):1319-30. doi: 10.1111/j.1365-2958.2004.04219.x. Mol Microbiol. 2004. PMID: 15387812 - Extensive surface diversity of a commensal microorganism by multiple DNA inversions.
Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. Krinos CM, et al. Nature. 2001 Nov 29;414(6863):555-8. doi: 10.1038/35107092. Nature. 2001. PMID: 11734857 - The Bacteroides fragilis cell envelope: quarterback, linebacker, coach-or all three?
Pumbwe L, Skilbeck CA, Wexler HM. Pumbwe L, et al. Anaerobe. 2006 Oct-Dec;12(5-6):211-20. doi: 10.1016/j.anaerobe.2006.09.004. Epub 2006 Oct 10. Anaerobe. 2006. PMID: 17045496 Review. - Site-specific inversion: enhancers, recombination proteins, and mechanism.
Craig NL. Craig NL. Cell. 1985 Jul;41(3):649-50. doi: 10.1016/s0092-8674(85)80040-4. Cell. 1985. PMID: 2988781 Review. No abstract available.
Cited by
- Analysis of a phase-variable restriction modification system of the human gut symbiont Bacteroides fragilis.
Ben-Assa N, Coyne MJ, Fomenkov A, Livny J, Robins WP, Muniesa M, Carey V, Carasso S, Gefen T, Jofre J, Roberts RJ, Comstock LE, Geva-Zatorsky N. Ben-Assa N, et al. Nucleic Acids Res. 2020 Nov 4;48(19):11040-11053. doi: 10.1093/nar/gkaa824. Nucleic Acids Res. 2020. PMID: 33045731 Free PMC article. - Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation.
Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. Kuwahara T, et al. Proc Natl Acad Sci U S A. 2004 Oct 12;101(41):14919-24. doi: 10.1073/pnas.0404172101. Epub 2004 Oct 4. Proc Natl Acad Sci U S A. 2004. PMID: 15466707 Free PMC article. - Characterization and induction of prophages in human gut-associated Bifidobacterium hosts.
Mavrich TN, Casey E, Oliveira J, Bottacini F, James K, Franz CMAP, Lugli GA, Neve H, Ventura M, Hatfull GF, Mahony J, van Sinderen D. Mavrich TN, et al. Sci Rep. 2018 Aug 24;8(1):12772. doi: 10.1038/s41598-018-31181-3. Sci Rep. 2018. PMID: 30143740 Free PMC article. - Phase-locked mutants of Mycoplasma agalactiae: defining the molecular switch of high-frequency Vpma antigenic variation.
Chopra-Dewasthaly R, Citti C, Glew MD, Zimmermann M, Rosengarten R, Jechlinger W. Chopra-Dewasthaly R, et al. Mol Microbiol. 2008 Mar;67(6):1196-210. doi: 10.1111/j.1365-2958.2007.06103.x. Epub 2008 Jan 30. Mol Microbiol. 2008. PMID: 18248580 Free PMC article. - Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem.
Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. Fletcher CM, et al. Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2413-8. doi: 10.1073/pnas.0608797104. Epub 2007 Feb 6. Proc Natl Acad Sci U S A. 2007. PMID: 17284602 Free PMC article.
References
- Bry, L., Falk, P. G., Midtvedt, T. & Gordon, J. I. (1996) Science 273, 1380-1383. - PubMed
- Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., Hooper, L. V. & Gordon, J. I. (2003) Science 299, 2074-2076. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources