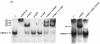Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells - PubMed (original) (raw)
Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells
Taka-aki Matsuoka et al. Mol Cell Biol. 2003 Sep.
Abstract
The C1/RIPE3b1 (-118/-107 bp) binding factor regulates pancreatic-beta-cell-specific and glucose-regulated transcription of the insulin gene. In the present study, the C1/RIPE3b1 activator from mouse beta TC-3 cell nuclear extracts was purified by DNA affinity chromatography and two-dimensional gel electrophoresis. C1/RIPE3b1 binding activity was found in the roughly 46-kDa fraction at pH 7.0 and pH 4.5, and each contained N- and C-terminal peptides to mouse MafA as determined by peptide mass mapping and tandem spectrometry. MafA was detected in the C1/RIPE3b1 binding complex by using MafA peptide-specific antisera. In addition, MafA was shown to bind within the enhancer region (-340/-91 bp) of the endogenous insulin gene in beta TC-3 cells in the chromatin immunoprecipitation assay. These results strongly suggested that MafA was the beta-cell-enriched component of the RIPE3b1 activator. However, reverse transcription-PCR analysis demonstrated that mouse islets express not only MafA but also other members of the large Maf family, specifically c-Maf and MafB. Furthermore, immunohistochemical studies revealed that at least MafA and MafB were present within the nuclei of islet beta cells and not within pancreas acinar cells. Because MafA, MafB, and c-Maf were each capable of specifically binding to and activating insulin C1 element-mediated expression, our results suggest that all of these factors play a role in islet beta-cell function.
Figures
FIG. 1.
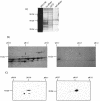
InsC1/RIPE3b1 binding activity localized to pH 7 and pH 4.5 after 2-D electrophoresis. (A) Southwestern analysis was performed by using 2-D electrophoresis resolved βTC-3 nuclear extract (NE). InsC1 binding of a 46-kDa protein(s) was found at roughly pH 7 and pH 4.5. (B) The proteins within the 46-kDa range were eluted from 2-D slices representing different pH ranges, renatured, and tested for InsC1 binding in the gel shift assay. The arrow marks the mobility of the RIPE3b1 complex in unfractionated, control βTC-3 nuclear extract (NE). (C) Gel shift reactions were conducted with the pH 7 fraction obtained from 2-D resolved βTC-3 nuclear extract. A 10-fold molar excess of unlabeled wild-type and RIPE3b1 binding-defective −111/−108 InsC1 competitor was used in gel shift reactions.
FIG. 2.
Biochemical isolation of the InsC1/RIPE3b1 DNA-binding subunit. (A) InsC1 affinity chromatography was performed on βTC-3 nuclear extract (NE). The protein composition of each step was determined by SDS-PAGE and silver staining. First affinity, InsC1-(AC)5 Sepharose chromatography; (AC)5, Sepharose chromatography alone; second affinity, InsC1-(AC)5 Sepharose chromatography. (B) The pooled InsC1/RIPE3b1 binding fractions from the second affinity step were subjected to 2-D electrophoresis. The Coomassie blue-stained spots at pH 7.0 and 4.5 (arrow) of approximately 46-kDa were subjected to MALDI-TOF mass spectrometry. (C) βTC-3 nuclear extract (NE) was resolved by 2-D electrophoresis and probed by Western analysis with αMafA.
FIG. 3.
Alignment of mouse MafA with other large Maf family members. The amino acid identity between mouse MafA, mouse MafB, mouse c-Maf, and mouse NRL is depicted by the gray shading. The underlined amino acids represent the eight tryptic peptides identified by mass spectrometry. All of these peptides were found in the pH 7 and pH 4.5 samples. Amino acids 232 to 324 of mouse MafA span the conserved basic leucine-zipper region involved in DNA binding and dimerization, whereas the less-characterized S/T-rich activation domain is likely found between amino acids 1 and 75 (4, 73).
FIG. 4.
MafA is found in the βTC-3 and human islet InsC1/RIPE3b1 activator complex. Gel shift binding to the InsC1 probe was conducted with βTC-3 (A) or human islet (B) nuclear extract in the absence (−) or presence of αMafA, αlarge Maf (Maf#153), αMafB, and/or αc-Maf antibody. The arrows represent the supershifted (SS) complexes. The more broadly recognizing Maf#153 antisera (see panel C) completely altered RIPE3b1 mobility, whereas αMafA affected a portion of the (A) βTC-3 nuclear extract activity. The αMafA supershift in panel A was blocked by the addition of the antigenic MafA332/342 peptide. αc-Maf and αMafB also affected RIPE3b1 in βTC-3 (A) or human islet (B) nuclear extract, respectively. The RIPE3b1 complex in panel B was identified by competition with wild-type InsC1 (lane W) and the −111/−108 bp binding mutant (lane M). (C) Nuclear extracts from MafA-, MafB-, c-Maf-, and pcDNA3.1 [lanes (−)]-transfected HeLa cells were analyzed with αMafA, αMafB, αc-Maf, and/or αlarge Maf antisera by Western analysis. The asterisk denotes the location of the large Maf product.
FIG. 4.
MafA is found in the βTC-3 and human islet InsC1/RIPE3b1 activator complex. Gel shift binding to the InsC1 probe was conducted with βTC-3 (A) or human islet (B) nuclear extract in the absence (−) or presence of αMafA, αlarge Maf (Maf#153), αMafB, and/or αc-Maf antibody. The arrows represent the supershifted (SS) complexes. The more broadly recognizing Maf#153 antisera (see panel C) completely altered RIPE3b1 mobility, whereas αMafA affected a portion of the (A) βTC-3 nuclear extract activity. The αMafA supershift in panel A was blocked by the addition of the antigenic MafA332/342 peptide. αc-Maf and αMafB also affected RIPE3b1 in βTC-3 (A) or human islet (B) nuclear extract, respectively. The RIPE3b1 complex in panel B was identified by competition with wild-type InsC1 (lane W) and the −111/−108 bp binding mutant (lane M). (C) Nuclear extracts from MafA-, MafB-, c-Maf-, and pcDNA3.1 [lanes (−)]-transfected HeLa cells were analyzed with αMafA, αMafB, αc-Maf, and/or αlarge Maf antisera by Western analysis. The asterisk denotes the location of the large Maf product.
FIG. 4.
MafA is found in the βTC-3 and human islet InsC1/RIPE3b1 activator complex. Gel shift binding to the InsC1 probe was conducted with βTC-3 (A) or human islet (B) nuclear extract in the absence (−) or presence of αMafA, αlarge Maf (Maf#153), αMafB, and/or αc-Maf antibody. The arrows represent the supershifted (SS) complexes. The more broadly recognizing Maf#153 antisera (see panel C) completely altered RIPE3b1 mobility, whereas αMafA affected a portion of the (A) βTC-3 nuclear extract activity. The αMafA supershift in panel A was blocked by the addition of the antigenic MafA332/342 peptide. αc-Maf and αMafB also affected RIPE3b1 in βTC-3 (A) or human islet (B) nuclear extract, respectively. The RIPE3b1 complex in panel B was identified by competition with wild-type InsC1 (lane W) and the −111/−108 bp binding mutant (lane M). (C) Nuclear extracts from MafA-, MafB-, c-Maf-, and pcDNA3.1 [lanes (−)]-transfected HeLa cells were analyzed with αMafA, αMafB, αc-Maf, and/or αlarge Maf antisera by Western analysis. The asterisk denotes the location of the large Maf product.
FIG. 5.
MafA is present in the RIPE3b1 activator complex immunoprecipitated by anti-phosphotyrosine antisera. (A) Schematic of the antiphosphotyrosine antibody immunoprecipitation procedure. (B) βTC-3 nuclear extract was immunoprecipitated with normal IgG (lane 1) or antiphosphotyrosine 4G10 monoclonal antibody (lanes 2 to 4). The immunoprecipitated material was washed, and the released proteins were electrotransferred from an SDS-PAGE gel onto an Immobilon polyvinylidene difluoride membrane. The proteins eluted from the roughly 46-kDa membrane slice were assayed for InsC1 binding activity in the absence or presence of αMafA (lane 3) or Maf#153 (lane 4) antibodies. The RIPE3b1 complex eluted directly after SDS-PAGE and Immobilon fractionation is also shown (lane 5). These results are representative of an experiment repeated on several separate occasions.
FIG. 6.
MafA binds within the enhancer region of the endogenous insulin gene. The cross-linked DNA immunoprecipitated from βTC-3 cells with αMafA antibody was analyzed by PCR with insulin (−378/−46) and PEPCK control region-specific primers (lane 3). As controls, PCRs were run with no DNA (lane 1), on input chromatin (1:100 dilution, lane 2), or with DNA obtained after precipitation with rabbit IgG (lane 4) or no antibody (lane 5). The same pattern was obtained with the Maf#153 antisera (data not shown).
FIG. 7.
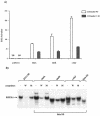
Islets express MafA, MafB, and c-Maf mRNA. (A) RT-PCR analysis was performed on RNA isolated from mouse islets, βTC3 cells, and αTC-6 cells with large Maf-specific primers. MafA, MafB, and c-Maf, but not NRL, were amplified to various degrees from each source. These RNAs were not detected without (− lanes) the addition of reverse transcriptase or in the absence of RNA. The correctness of the RT-PCR products was determined by DNA sequencing and by amplification of the same size PCR product from the intronless primer spanning sequences by using mouse genomic DNA. (B) Determination of MafA, MafB, and c-Maf levels in the adult islet by quantitative competitive RT-PCR. Increasing amounts of the large Maf DNA competitor were added to the RT-PCRs. The arrow denotes the location of the competitor band. The relative levels of MafA, MafB, and c-Maf were determined by densitometric quantification and correspond to approximately 1.0, 0.5, and 0.01, respectively.
FIG. 8.
MafA, MafB, and c-Maf stimulate InsC1-driven transcription. (A) HeLa cells were transfected with the wild-type (WT) or the RIPE3b1 binding defective −111/−108 InsC1 mutant (M) of the insulin-driven −238 LUC expression plasmid, the pRL-CMV internal transfection control, and MafA, MafB, and c-Maf subcloned into the CMV-driven pcDNA3.1 expression plasmid. The Maf activation signal is only partially reduced in the −238-Insulin C1 mutant due to the presence of additional sites in this region of the rat insulin II gene at −152/−140 bp and −94/−82 bp (data not shown). The data are presented as the large Maf transfected mean activity ± the standard error relative to the pcDNA3.1 alone from at least four separate experiments. The relative light unit activities of −238-Insulin and −238-Insulin C1 M alone were 13,361 ± 3,320 and 18,877 ± 4,258, respectively. (B) Competition analysis performed with nuclear extracts (NEs) prepared from MafA, MafB, and c-Maf transfected HeLa cells demonstrates RIPE3b1-like gel shift binding of the transfected proteins to the InsC1 wild type (lanes W) and −111/-108 bp mutant (lanes M). The c-Maf binding reactions contain three times more nuclear extract (6 μg) than did the MafA or MafB samples. No detectable
l-
Maf-binding activity was found in the untransfected HeLa nuclear extract.
FIG. 9.
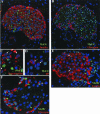
MafA and MafB are localized in the nuclei of islet β cells. Immunohistochemical detection of MafA (A to D) and MafB (E and F) expression in adult mouse pancreas. Double immunofluorescence was used to analyze coexpression of these factors with hormones (red): insulin (A and E), glucagon (B and F), somatostatin (C), and/or PP (D). The nuclei are counterstained in blue. Note that MafA staining (green) was only detected in insulin-producing cells, whereas MafB was expressed in both insulin- and glucagon-producing cells. In addition, MafA and MafB immunoreactivity was not observed in surrounding exocrine cells. The arrows in panels C and D denote the absence of MafA staining in δ and PP cells and the presence of MafB expression in β (E) and α (F) cells. The c-Maf-specific antiserum (N-15) did not stain control tissues, preventing the determination of c-Maf expression in islets (data not shown).
Similar articles
- The islet beta cell-enriched RIPE3b1/Maf transcription factor regulates pdx-1 expression.
Samaras SE, Zhao L, Means A, Henderson E, Matsuoka TA, Stein R. Samaras SE, et al. J Biol Chem. 2003 Apr 4;278(14):12263-70. doi: 10.1074/jbc.M210801200. Epub 2003 Jan 27. J Biol Chem. 2003. PMID: 12551916 - MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene.
Kataoka K, Han SI, Shioda S, Hirai M, Nishizawa M, Handa H. Kataoka K, et al. J Biol Chem. 2002 Dec 20;277(51):49903-10. doi: 10.1074/jbc.M206796200. Epub 2002 Oct 3. J Biol Chem. 2002. PMID: 12368292 - Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA.
Olbrot M, Rud J, Moss LG, Sharma A. Olbrot M, et al. Proc Natl Acad Sci U S A. 2002 May 14;99(10):6737-42. doi: 10.1073/pnas.102168499. Proc Natl Acad Sci U S A. 2002. PMID: 12011435 Free PMC article. - Roles and regulation of transcription factor MafA in islet beta-cells.
Aramata S, Han SI, Kataoka K. Aramata S, et al. Endocr J. 2007 Dec;54(5):659-66. doi: 10.1507/endocrj.kr-101. Epub 2007 Aug 30. Endocr J. 2007. PMID: 17785922 Review. - PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration.
Zhu Y, Liu Q, Zhou Z, Ikeda Y. Zhu Y, et al. Stem Cell Res Ther. 2017 Nov 2;8(1):240. doi: 10.1186/s13287-017-0694-z. Stem Cell Res Ther. 2017. PMID: 29096722 Free PMC article. Review.
Cited by
- Cooperation between HMGA1, PDX-1, and MafA is Essential for Glucose-Induced Insulin Transcription in Pancreatic Beta Cells.
Arcidiacono B, Iiritano S, Chiefari E, Brunetti FS, Gu G, Foti DP, Brunetti A. Arcidiacono B, et al. Front Endocrinol (Lausanne). 2015 Jan 13;5:237. doi: 10.3389/fendo.2014.00237. eCollection 2014. Front Endocrinol (Lausanne). 2015. PMID: 25628604 Free PMC article. - Purification and characterization of transcription factors.
Nagore LI, Nadeau RJ, Guo Q, Jadhav YL, Jarrett HW, Haskins WE. Nagore LI, et al. Mass Spectrom Rev. 2013 Sep-Oct;32(5):386-98. doi: 10.1002/mas.21369. Epub 2013 Jul 7. Mass Spectrom Rev. 2013. PMID: 23832591 Free PMC article. Review. - Reprogramming into pancreatic endocrine cells based on developmental cues.
Kordowich S, Mansouri A, Collombat P. Kordowich S, et al. Mol Cell Endocrinol. 2010 Feb 5;315(1-2):11-8. doi: 10.1016/j.mce.2009.10.015. Epub 2009 Nov 6. Mol Cell Endocrinol. 2010. PMID: 19897012 Free PMC article. Review. - Pancreatic islet cell therapy for type I diabetes: understanding the effects of glucose stimulation on islets in order to produce better islets for transplantation.
Ren J, Jin P, Wang E, Liu E, Harlan DM, Li X, Stroncek DF. Ren J, et al. J Transl Med. 2007 Jan 3;5:1. doi: 10.1186/1479-5876-5-1. J Transl Med. 2007. PMID: 17201925 Free PMC article. Review. - Effect of PTTG on endogenous gene expression in HEK 293 cells.
Panguluri SK, Kakar SS. Panguluri SK, et al. BMC Genomics. 2009 Dec 3;10:577. doi: 10.1186/1471-2164-10-577. BMC Genomics. 2009. PMID: 19958546 Free PMC article.
References
- Benkhelifa, S., S. Provot, O. Lecoq, C. Pouponnot, G. Calothy, and M. P. Felder-Schmittbuhl. 1998. mafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene 17:247-254. - PubMed
- Blank, V., and N. C. Andrews. 1997. The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci. 22:437-441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P60 DK020593/DK/NIDDK NIH HHS/United States
- P01 DK42502/DK/NIDDK NIH HHS/United States
- R01 GM043609/GM/NIGMS NIH HHS/United States
- GM43609/GM/NIGMS NIH HHS/United States
- P60 DK20593/DK/NIDDK NIH HHS/United States
- P01 DK042502/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases