BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation - PubMed (original) (raw)
BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation
Pei-Wen Hsiao et al. Mol Cell Biol. 2003 Sep.
Abstract
Nuclear hormone receptors are ligand-dependent transcriptional regulators that modulate chromatin structure. However, the precise molecular mechanisms by which receptors recruit chromatin-remodeling activity are not fully elucidated. We show that in the absence of its ligand-binding domain, the glucocorticoid receptor (GR) is able to interact with both nuclear receptor coactivators and the BRG1 chromatin-remodeling complex in vivo. Individually, the GR makes direct interactions with BRG1-associated factor 60a (BAF60a) and BAF57, but not with BRG1, BAF155, or BAF170. Further, BAF60a possesses at least two interaction surfaces, one for GR and BRG1 and a second for BAF155 and BAF170. A GR mutant, GR(R488Q), that fails to interact with BAF60a in vitro has reduced chromatin-remodeling activity and reduced transcriptional activity from the promoter assembled as chromatin in vivo. Stable expression of a BAF60a truncation mutant, BAF60a4-140, caused chromatin-specific loss of GR functions in vivo. In the presence of the BAF60a mutant, the GR fails to interact with the BRG1 complex and consequently is also deficient in its ability to activate transcription from chromatin. Thus, in addition to previously identified BAF250, BAF60a may provide another critical and direct link between nuclear receptors and the BRG1 complex that is required for promoter recruitment and subsequent chromatin remodeling.
Figures
FIG. 1.
GR1-556 interacts with SRC-1 and BRG1 complex in vivo. (A) 2963.1 cells, which express endogenous PR, and derived 2963.1/556 cells were treated with ethanol (−), dexamethasone (Dex) (10−7 M), or R5020 (10−8 M) for 24 h. The CAT activity of cell lysate was analyzed by kinetic assays and normalized with total protein. The insert shows expression of GR1-556 in 2963.1/556 cells but not in the parental 2963.1 cells or A1-2 cells which express the GRwt. (B) SRC-1 associates with GR1-556 in 2963.1/556 cells. GR1-556-associated complexes were immunoprecipitated from whole-cell extracts of 2963.1/556 cells with no antibody (lane 3) or BUGR2 anti-GR antibody (lane 4) and immunoblotted with antibodies against SRC-1, BRG1, BAF155, and GR (BUGR2). The cognate inputs of SRC-1, BRG1, and BAF155 are shown in lanes 1 and 2 for the no-antibody (No Ab) and anti-GR antibody (αGR) immunoblots, respectively. (C) Interaction of GR1-556 with SRC-1, SRC-3, BRG1, BAF170, BAF155, BAF60a, and BAF57. GST and GR fusion proteins were used to pull down SRC-1, SRC-3, HDAC1, BRG1, and BAF170, 155, 60a, or 57. Input lysate (10%) was loaded as control.
FIG. 2.
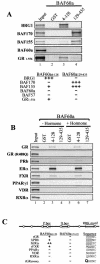
BAF60a links steroid hormone receptors and the BRG1 complex. (A) GST and BAF60a fusion proteins were used to pull down BRG1, BAF170, BAF155, BAF60a, and GR1-556, as described in the legend for Fig. 1C. The relative strengths of the interactions are indicated below the autoradiogram. (B) In vitro pull-down assays de-scribed in the legend for panel A were performed with GR, mutant GR(R488Q), PRb, ERα, FXR, PPARγ1, VDR, and RXRα. Hormones were applied at 1 μM dexamethasone for GR and 10 nM R5020 (PRb), 0.1 μM 17β-estradiol (ERα), 10 μM chenodeoxycholic acid (FXR), 10 μM 15d-prostaglandin J2 (PPARγ1), 10 nM EB1089 (VDR), or 10 μM 9-cis retinoic acid (RXRα) and compared to results for the vehicle without hormone (− Hormone). (C) Sequence alignment of the nuclear receptors used in the pull-down assays to illustrate the RRK motif in the DBD.
FIG. 3.
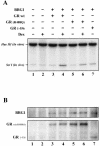
GR(R488Q) mutant fails to interact with BAF60a in vitro and is unable to activate transcription from a chromatin template in vivo. (A) GRwt and GR(R488Q) were transfected with BRG1 and MMTV-CAT reporter into SW-13/M3-2 cells. Cells were treated posttransfection with ethanol (−) or dexamethasone (10−7 M) (+) for 24 h. CAT and luciferase activities in the same cell lysate were plotted according to dexamethasone induction. (B) Expression of transfected BRG1, GRwt, and GR(R488Q) was examined by Western blotting.
FIG. 4.
Both GR1-556 and the GR(R488Q) mutant exhibit decreased chromatin remodeling. (A) Chromatin remodeling stimulated by GRwt, GR(R488Q), or GR1-556 was examined by _Sst_I hypersensitivity. SW-13/M3-2 cells were transfected with BRG1 and GRwt, GR(R488Q), or GR1-556 for 24 h. Nuclei were isolated from the transfected cells after treatment with dexamethasone (10−7 M) (+) or control (ethanol) (−) and were immediately subjected to limited digestion by _Sst_I to assess hypersensitivity of Nuc-B within the chromatin promoter. (B) The BRG1 complex interaction with GRwt, GR(R488Q), or GR1-556 in transfected SW-13/M3-2 cells was examined by coimmunoprecipitation with BRG1 antibody from 500 μg of whole-cell extract followed by Western blot analysis with antibodies against BRG1 and GR.
FIG. 5.
Expression of BAF60a4-140 blocks GR activation. (A) Upper panel, UL3 cells and derived BAF60a4-140-expressing clones 60N.9, 60N.15, and 60N.17 were treated with ethanol (−) or 10−8 M dexamethasone (+) for 24 h. Chromatin-assembled MMTV-luciferase expression was plotted as the percentage of dexamethasone (Dex)-induced activity in UL3 cells. Lower panel, BAF60a and BAF60a4-140 were detected in nuclear extracts from UL3 cells and BAF60a4-140-derived clones by immunoblotting with antibody against BAF60a4-64. (B) Expression of BAF60a4-140 inhibits GR activation of the chromatin template, but not the transient template. An MMTV-CAT plasmid was transfected into UL3 and 60N.17 cells. Cells were treated posttransfection with dexamethasone (10−8 M) (+) or ethanol (−) for 24 h. CAT and luciferase activities in the same cell lysate were plotted using the dexamethasone-induced activity of UL3 cells as 100%. (C) Chromatin structure of the MMTV promoter in UL3 and 60N.17 cells was examined by _Sst_I hypersensitivity after treatment with ethanol (−) or 10−8 M dexamethasone (+) for 1 h. The _Sst_I hypersensitivity of each lane is plotted relative to that of lane 1 from the right panel. Error bars represent standard deviations of three trials.
FIG. 6.
BAF60a4-140 inhibits GR interaction with the BRG1 complex. (A) GR interaction with the BRG1 complex was examined by coimmunoprecipitation with an anti-BRG1 antibody. Input (100 μg of whole-cell extract) and the coimmunoprecipitated complex (from 1 mg of whole-cell extract) were analyzed by Western blotting with antibodies against BRG1, BAF155, GR, and BAF60a. (B) Models for BAF60a4-140 inhibition of GR interaction with the BRG1 complex. The BAF60a4-140 mutant may integrate into BRG1 complex alone or together with BAF60a to generate a nonfunctional complex. Alternatively, the BAF60a4-140 mutant may directly block the ability of the GR to interact with the complex.
FIG. 6.
BAF60a4-140 inhibits GR interaction with the BRG1 complex. (A) GR interaction with the BRG1 complex was examined by coimmunoprecipitation with an anti-BRG1 antibody. Input (100 μg of whole-cell extract) and the coimmunoprecipitated complex (from 1 mg of whole-cell extract) were analyzed by Western blotting with antibodies against BRG1, BAF155, GR, and BAF60a. (B) Models for BAF60a4-140 inhibition of GR interaction with the BRG1 complex. The BAF60a4-140 mutant may integrate into BRG1 complex alone or together with BAF60a to generate a nonfunctional complex. Alternatively, the BAF60a4-140 mutant may directly block the ability of the GR to interact with the complex.
Similar articles
- BAF60A mediates interactions between the microphthalmia-associated transcription factor and the BRG1-containing SWI/SNF complex during melanocyte differentiation.
Aras S, Saladi SV, Basuroy T, Marathe HG, Lorès P, de la Serna IL. Aras S, et al. J Cell Physiol. 2019 Jul;234(7):11780-11791. doi: 10.1002/jcp.27840. Epub 2018 Dec 4. J Cell Physiol. 2019. PMID: 30515787 Free PMC article. - Reconstitution of glucocorticoid receptor-dependent transcription in vivo.
Trotter KW, Archer TK. Trotter KW, et al. Mol Cell Biol. 2004 Apr;24(8):3347-58. doi: 10.1128/MCB.24.8.3347-3358.2004. Mol Cell Biol. 2004. PMID: 15060156 Free PMC article. - The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo.
Trotter KW, Fan HY, Ivey ML, Kingston RE, Archer TK. Trotter KW, et al. Mol Cell Biol. 2008 Feb;28(4):1413-26. doi: 10.1128/MCB.01301-07. Epub 2007 Dec 17. Mol Cell Biol. 2008. PMID: 18086889 Free PMC article. - Chromatin remodeling during glucocorticoid receptor regulated transactivation.
King HA, Trotter KW, Archer TK. King HA, et al. Biochim Biophys Acta. 2012 Jul;1819(7):716-26. doi: 10.1016/j.bbagrm.2012.02.019. Epub 2012 Mar 6. Biochim Biophys Acta. 2012. PMID: 22425674 Free PMC article. Review. - The BRG1 transcriptional coregulator.
Trotter KW, Archer TK. Trotter KW, et al. Nucl Recept Signal. 2008 Feb 1;6:e004. doi: 10.1621/nrs.06004. Nucl Recept Signal. 2008. PMID: 18301784 Free PMC article. Review.
Cited by
- SWI/SNF Complex in Vascular Smooth Muscle Cells and Its Implications in Cardiovascular Pathologies.
Liu H, Zhao Y, Zhao G, Deng Y, Chen YE, Zhang J. Liu H, et al. Cells. 2024 Jan 16;13(2):168. doi: 10.3390/cells13020168. Cells. 2024. PMID: 38247859 Free PMC article. Review. - DNA and chromatin modification networks distinguish stem cell pluripotent ground states.
Song J, Saha S, Gokulrangan G, Tesar PJ, Ewing RM. Song J, et al. Mol Cell Proteomics. 2012 Oct;11(10):1036-47. doi: 10.1074/mcp.M111.011114. Epub 2012 Jul 22. Mol Cell Proteomics. 2012. PMID: 22822199 Free PMC article. - Mechanism of action of the SWI/SNF family complexes.
Chen K, Yuan J, Sia Y, Chen Z. Chen K, et al. Nucleus. 2023 Dec;14(1):2165604. doi: 10.1080/19491034.2023.2165604. Nucleus. 2023. PMID: 36633435 Free PMC article. No abstract available. - Combinatorial probabilistic chromatin interactions produce transcriptional heterogeneity.
Voss TC, Schiltz RL, Sung MH, Johnson TA, John S, Hager GL. Voss TC, et al. J Cell Sci. 2009 Feb 1;122(Pt 3):345-56. doi: 10.1242/jcs.035865. Epub 2009 Jan 6. J Cell Sci. 2009. PMID: 19126674 Free PMC article. - Role of brahma-related gene 1/brahma-associated factor subunits in neural stem/progenitor cells and related neural developmental disorders.
Ke NY, Zhao TY, Wang WR, Qian YT, Liu C. Ke NY, et al. World J Stem Cells. 2023 Apr 26;15(4):235-247. doi: 10.4252/wjsc.v15.i4.235. World J Stem Cells. 2023. PMID: 37181007 Free PMC article. Review.
References
- Archer, T. K., P. Lefebvre, R. G. Wolford, and G. L. Hager. 1992. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science 255:1573-1576. - PubMed
- Archer, T. K., E. Zaniewski, M. L. Moyer, and S. K. Nordeen. 1994. The differential capacity of glucocorticoids and progestins to alter chromatin structure and induce gene expression in human breast cancer cells. Mol. Endocrinol. 8:1154-1162. - PubMed
- Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous