GATA-3: an unexpected regulator of cell lineage determination in skin - PubMed (original) (raw)
. 2003 Sep 1;17(17):2108-22.
doi: 10.1101/gad.1115203. Epub 2003 Aug 15.
Affiliations
- PMID: 12923059
- PMCID: PMC196453
- DOI: 10.1101/gad.1115203
GATA-3: an unexpected regulator of cell lineage determination in skin
Charles K Kaufman et al. Genes Dev. 2003.
Abstract
Multipotent skin stem cells give rise to epidermis and its appendages, including the hair follicle. The Lef-1/Tcf family of Wnt-regulated transcription factors plays a major role in specification of the hair shaft, but little is known about how the equally important hair channel, the inner root sheath (IRS), develops in concert to shape and guide the hair. In a microarray screen to search for transcriptional regulators of hair follicle morphogenesis, we identified GATA-3, a key regulator of T-cell lineage determination. Surprisingly, this transcription factor is essential for stem cell lineage determination in skin, where it is expressed at the onset of epidermal stratification and IRS specification in follicles. GATA-3-null/lacZ knock-in embryos can survive up to embryonic day 18.5 (E18.5), when they fail to form the IRS. Skin grafting unveiled additional defects in GATA-3-null hairs and follicles. IRS progenitors failed to differentiate, whereas cortical progenitors differentiated, but produced an aberrant hair structure. Curiously, some GATA-3-null progenitor cells expressed mixed IRS and hair shaft markers. Taken together, these findings place GATA-3 with Lef-1/Wnts at the crossroads of the IRS versus hair shaft cell fate decision in hair follicle morphogenesis. This newfound function for GATA-3 in skin development strengthens the parallels between the differentiation programs governing hair follicle and lymphocyte differentiation.
Figures
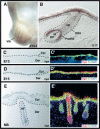
Figure 1.
GATA-3 is expressed in embryonic skin epithelium. (A,B) Whole mount in situ hybridization with a GATA-3 digoxygenin cRNA probe (brown). (A) Whole embryo hybridization, depicting GATA-3 mRNA in developing vibrissae (Vib) at E14.5. (B) Frozen section of skin from E17 whole mount in situ hybridization. Note strong hybridization in suprabasal epidermis and progenitor cells for the IRS (preIRS). (C_–_E) Frozen sections of GATA-3nlslacZ/+ embryos, at ages indicated in lower left of each frame. Sections were subjected to either X-gal staining (C,D,E) or immunofluorescence (_C_′, _D_′, _E_′) with DAPI to detect nuclei, and with antibodies (Abs) against β-galactosidase (Bgal) to detect GATA-3 promoter activity and against K5 to detect the basal epidermal layer and follicle ORS. Abs are color-coded in the lower right of each frame. (Epi) Epidermis; (Der) dermis; (HF) hair follicle; (DP) dermal papilla; (ORS) outer root sheath; (M) matrix.
Figure 2.
GATA-3 expression correlates with IRS differentiation. Dorsal skins were from GATA-3nlslacz/+ mice at the postnatal days indicated in the_upper right_ of each frame. Skins were frozen, sectioned, and stained with DAPI and X-gal (A,B), or subjected to indirect triple immunofluorescence (C_–_E) with DAPI and Abs against β-galactosidase (Bgal) to detect GATA-3 promoter activity; AE13 to detect cortical acidic hair keratins; AE15 to detect IRS and medulla trichohyalin; K6 to detect the companion layer (Cp) keratins and the medulla K6 keratins; or GATA-3. Antibodies are color-coded as indicated on each frame. The stages of the hair cycle are as follows: P16, late anagen, first cycle; P21, early anagen, second cycle; P6, early to mid-anagen, first cycle. (A,B) 20× magnification, 10-μm sections. (C_–_E) Composites of two microscopic fields at 63× magnification, 6-μm sections. (ORS) outer root sheath; (Cp) companion cell layer; (IRS) inner root sheath; (He) Henle's layer; (Hu) Huxley's layer; (Ci) cuticle of IRS; (Ch) cuticle of hair shaft; (Co) cortex of hair shaft; (Me) medulla; (DP) dermal papilla. The diagram at_right_ summarizes the immunofluorescence patterns observed within the bulb of the hair follicle. The K6 antibody we use here does not recognize the IRS-specific K6 keratins.
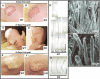
Figure 3.
Whisker follicles develop by E18.5, but they are aberrant in_GATA-3_-null embryos. Whisker pads of E18.5 wild-type (WT) and_GATA-3nlslacz(z/z)_ (GATA-3_-null) embryos were frozen and sectioned (10 μm). Serial sections (denoted by i, ii, iii) of two wild-type follicles (A,B) and two KO follicles (C,D) are shown. Sections were either subjected to H&E staining (A_–_D) or triple indirect immunofluorescence microscopy using the antibodies indicated in the lower right of each frame. Color coding reflects the secondary Abs used for detection. Brackets in_Ai and Bi denote a gap between the hair cortex (red, AE13) and companion cell layer (green, K6). This gap represents the IRS (red, AE15;Aii and Bii), which is sandwiched between the companion layer and the cortex of wild-type follicles. In KO follicles, AE15 staining is largely absent, and K6 and AE13 staining are adjacent to one another. Additional abnormalities in embryonic KO follicles include nodular thickenings in the vibrissae shafts (marked with an asterisk; C), irregular bulges in the ORS (e.g., double-headed arrow; D), and a shortened, bent follicle of enlarged diameter at the bulb. Images are shown at 20× magnification. Panels are composites of two or three microscopic fields. (E) Barrier function analyses. The ability of whole embryos to exclude blue dye was used to examine the epidermal barrier, normally acquired beginning at E17.5 and complete by E18.5. Note the complete penetration of blue dye in a representative _GATA-3_-null E17.5 embryo, in contrast to the comparably sized E17.5 wild-type embryo, which has the characteristic dorsal pattern of dye exclusion. By E18.5, both wild-type and null embryos have acquired barrier function based on this assay. Note, however, that whiskers have not protruded from the muzzle of the KO embryo, in contrast to the wild-type littermate.
Figure 4.
Classical morphological and biochemical features of epidermal differentiation are normal in _GATA-3_-null animals. Panels are from an E18.5 wild-type littermate (left set) and a _GATA-3_-null embryo (right set). (A,B) Dorsal back skins were fixed in Epon and embedded, and semithin sections (1 μm) were stained with toluidine blue. Note the presence of basal (BL), spinous (SL), granular (GL), and cornified (CL) layers, which are largely similar between wild-type and KO skin. Note also similarities in wild-type and KO pelage hair follicles (HF), which at this stage have not begun to differentiate into shaft, IRS, and ORS. (C_–_J) Frozen sections of E18.5 dorsal back skins were subjected to triple indirect immunofluorescence using DAPI to label the nuclei and the following antibodies: K5 to label the epidermal basal layer; K1 and involucrin (Inv; in mouse skin, Inv is more suprabasal than in human) to label the spinous layers; filaggrin (Fil) and loricrin (Lor) to mark the granular layers (Fil detects pre- and processed filaggrin). Similar normal biochemical patterns of staining were observed for mature epidermis, obtained from grafted KO and wild-type skins.
Figure 5.
_GATA-3_-null skin grafts develop strikingly abnormal hairs. Full thickness skins from six wild-type and six _GATA-3_-null (KO) E17.5 CD-1 albino embryos were grafted onto nu/nu mice. At 9 d after engraftment, hairs (white) appeared in all wild-type grafts (A) but were absent in all GATA-3_-null grafts (B). By 27–29 d, a thick pelage hair coat had developed on all wild-type grafts (C and_E show representative examples of grafts from skins of two different wild-type animals), but the KO grafts produced only short, stubby hairs (D and F show representative examples of grafts from skins of two different KO animals). The three major hair types (awl, guard, and zig-zag) were evident among hairs plucked from wild-type grafts (examples shown in G) at 12 wk postengraftment. In contrast, hairs from equivalent GATA-3_-null grafts (representative examples shown in_H) did not fit these classifications and were shorter and thicker than wild-type shafts. (I,J) Scanning electron microscopy (SEM) analysis. SEM (600×) of the surface of wild-type and KO grafted skin at 12 wk postengraftment. The outer surface of each hair shaft is the cuticle layer, which is highly ordered in the wild-type but not KO mice. Note additional irregularities in the shafts, reflecting in detail the differences noted at lower magnification.
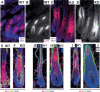
Figure 6.
Mature _GATA-3_-null hair follicles exhibit biochemical signs of an expanded cortex and medulla and a severely reduced inner root sheath. (A_–_J) Matched P35 skin grafts from E17.5_GATA-3_-null (KO) and wild-type littermates were frozen and sectioned (10 μm) and subjected to triple indirect immunofluorescence microscopy. Most follicles were in the anagen phase of the hair cycle. The upper right denotes genotype of skin. The antibodies used are noted on each frame and are color-coded according to the secondary Ab used for detection. DAPI was used to mark the nuclei, AE13 for cortical keratins, AE15 for IRS and medulla trichoyalin, anti-K6 for the companion layer and medulla (this antibody does not detect the IRS-K6 keratins), and Lef-1 to mark the precortical cells. denotes the wide stripe of AE13-negative, AE15-positive cells, corresponding to the medulla at the hair shaft center. Note the expanded Lef-1-positive precortex and AE13-positive cortex in mutant follicles. Brackets denote the AE15-positive IRS, largely absent in the KO follicles. Arrows indicate the lowest extent of AE15-positive IRS staining.
Figure 7.
Expansion and aberrancies in expression patterns of cortical and IRS precursor populations in _GATA-3_-null follicles. Skins from matched P35 wild-type and GATA-3_-null grafts were frozen, sectioned (10 μm), and subjected to either triple indirect immunofluorescence (A_–_K) or in situ hybridization (L,M). The genotype of each section is denoted at the upper right. Abs used are color-coded and indicated across the bottom of each frame. Asterisks in B and I denote autofluorescence, an artifact of the knots of highly keratinized material in some KO shafts. Boxed areas are shown at higher magnification in the insets. DAPI (blue) denotes nuclei. Abs were against GATA-3, present in the IRS lineage, and absent in the_GATA-3_-null sections; β-galactosidase (Bgal), specific for_GATA-3 promoter activity, which was indistinguishable from the anti-GATA IRS labeling in wild-type follicles, and still present in the expanded IRS precursor population of KO follicles; AE13, specific for the keratins of differentiating cortical cells; FOG1, a corepressor for GATA-3 protein, present in IRS precursor cells, where it marked the nuclei of wild-type but the cytoplasm of KO cells (D and E, respectively; see inset to E as well); Lef-1, specific for cortical precursor cells, expanded in the KO; Ki67, specific for proliferating cell nuclei; and K5, specific for the ORS of the follicle. The cRNA probe is against Shh, Sonic hedgehog, which marks a small subset of matrix cells in wild-type follicles, unchanged in KO follicles. Note that in KO follicles, some AE13-positive cortical and Lef-1-positive precortical cells were also positive for GATA-3 promoter activity (representative cells indicated by arrows in C and G). Note also that Ki67 labeling confirms the anagen state of both wild-type and KO follicles, and some Ki67-positive cells were also positive for GATA-3 promoter activity (J,K).
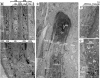
Figure 8.
Ultrastructural abnormalities in _GATA-3_-null follicles. Matched wild-type and _GATA-3_-null (KO) skin grafts were frozen in OCT (A,B) or fixed, embedded in Epon, and sectioned for ultrathin analyses (C,D). (A) Light microscopic analysis of H&E-stained 10-μm sections of 5-wk grafts. Wild-type (WT) composite of two 20× microscopic fields; KO is visible in one field because of its shorter and fatter structure at all stages of the hair cycle. (_B_–_C_″) Ultrastructural analyses of 12-wk grafts. All follicles are oriented with the skin surface at the top. Magnification bars are denoted on each frame.
Figure 8.
Ultrastructural abnormalities in _GATA-3_-null follicles. Matched wild-type and GATA-3_-null (KO) skin grafts were frozen in OCT (A,B) or fixed, embedded in Epon, and sectioned for ultrathin analyses (C,D). (B) Sagittal section of wild-type follicle, depicting the thin Henle's cell layer (He) with uniform density caused by keratinization. On the outside, this layer is flanked by the companion cell layer (Cp) and ORS and finally the dermal sheath (DS). On the inside, the Henle's layer is flanked by the trichohyalin (Th)-rich Huxley's (Hu) layer of the IRS, and the thin IRS cuticle (Ci). The hair shaft (HS) is internal to the IRS, and is composed of three major layers: an inner core of medulla (Me) cells with trichoyalin granules below each nucleus (arrowheads), the cortex (Co), and the cuticle of the hair shaft (Ch). The upper and_lower panels are from the same hair follicle, but the lower panel depicts the base of the keratinized Henle's layer of the IRS (double arrowheads); cells below this juncture still possess trichoyalin granules, but are less mature and less organized, as is characteristic of IRS precursor cells. (_C_–_C_″) Sections from _GATA-3_-null follicles. All are views from the region just above the follicle bulb. (C) The origins of the keratinized cells of a Henle's-like layer in a KO follicle (double arrows, IRS*). Note the presence of only a few trichoyalin granules (arrowheads) in the IRS precursor cells just below these keratinizing cells, in contrast to the wild-type IRS precursors, which are abundant in trichohyalin (Th; lower panel in B). Note also the presence of precortical cells (PreCo), rich in keratin filaments (Kf), located atypically below the Henle's cells; this irregularity was predicted from our biochemical studies (see Fig. 6F). Note further the mass of highly keratinized cortex and medulla central to the tiny IRS. The sections in _C_′ and_C_″ show that in some areas, the IRS was missing altogether, and the ORS and companion layer were abutted against the precortex, cortex, and medulla. Again, note signs of gross disorganization, with as many as three medulla cells (Me in _C_″) stacked horizontally against each other, instead of in the organized linear vertical array seen in wild-type medulla (B, upper panel). Note trichohyalin (arrowheads) in the medulla but not elsewhere (_C_″).
Similar articles
- Defining BMP functions in the hair follicle by conditional ablation of BMP receptor IA.
Kobielak K, Pasolli HA, Alonso L, Polak L, Fuchs E. Kobielak K, et al. J Cell Biol. 2003 Nov 10;163(3):609-23. doi: 10.1083/jcb.200309042. J Cell Biol. 2003. PMID: 14610062 Free PMC article. - Expression of GATA-3 in epidermis and hair follicle: relationship to p63.
Chikh A, Sayan E, Thibaut S, Lena AM, DiGiorgi S, Bernard BA, Melino G, Candi E. Chikh A, et al. Biochem Biophys Res Commun. 2007 Sep 14;361(1):1-6. doi: 10.1016/j.bbrc.2007.06.069. Epub 2007 Jul 2. Biochem Biophys Res Commun. 2007. PMID: 17632082 - Expression pattern of GATA-3 in embryonic and fetal human skin suggests a role in epidermal and follicular morphogenesis.
Sellheyer K, Krahl D. Sellheyer K, et al. J Cutan Pathol. 2010 Mar;37(3):357-61. doi: 10.1111/j.1600-0560.2009.01416.x. Epub 2009 Aug 27. J Cutan Pathol. 2010. PMID: 19719829 - Beauty is skin deep: the fascinating biology of the epidermis and its appendages.
Fuchs E. Fuchs E. Harvey Lect. 1998-1999;94:47-77. Harvey Lect. 1998. PMID: 11070952 Review. - GATA transcription factors and fat cell formation.
Tong Q, Tsai J, Hotamisligil GS. Tong Q, et al. Drug News Perspect. 2003 Nov;16(9):585-8. doi: 10.1358/dnp.2003.16.9.829340. Drug News Perspect. 2003. PMID: 14702139 Review.
Cited by
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by miR-22.
Yuan S, Li F, Meng Q, Zhao Y, Chen L, Zhang H, Xue L, Zhang X, Lengner C, Yu Z. Yuan S, et al. PLoS Genet. 2015 May 28;11(5):e1005253. doi: 10.1371/journal.pgen.1005253. eCollection 2015 May. PLoS Genet. 2015. PMID: 26020521 Free PMC article. - Scalp metastasis from occult primary breast carcinoma: A case report and review of the literature.
Alizadeh N, Mirpour H, Azimi SZ. Alizadeh N, et al. Int J Womens Dermatol. 2018 Sep 8;4(4):230-235. doi: 10.1016/j.ijwd.2018.08.002. eCollection 2018 Dec. Int J Womens Dermatol. 2018. PMID: 30627623 Free PMC article. - Mice with a targeted mutation of patched2 are viable but develop alopecia and epidermal hyperplasia.
Nieuwenhuis E, Motoyama J, Barnfield PC, Yoshikawa Y, Zhang X, Mo R, Crackower MA, Hui CC. Nieuwenhuis E, et al. Mol Cell Biol. 2006 Sep;26(17):6609-22. doi: 10.1128/MCB.00295-06. Mol Cell Biol. 2006. PMID: 16914743 Free PMC article. - GATA3 is downregulated in osteosarcoma and facilitates EMT as well as migration through regulation of slug.
Ma L, Xue W, Ma X. Ma L, et al. Onco Targets Ther. 2018 Oct 30;11:7579-7589. doi: 10.2147/OTT.S176534. eCollection 2018. Onco Targets Ther. 2018. PMID: 30464506 Free PMC article. - GATA3 Truncating Mutations Promote Cistromic Re-Programming In Vitro, but Not Mammary Tumor Formation in Mice.
Cornelissen LM, de Bruijn R, Henneman L, Kim Y, Zwart W, Jonkers J. Cornelissen LM, et al. J Mammary Gland Biol Neoplasia. 2019 Sep;24(3):271-284. doi: 10.1007/s10911-019-09432-4. Epub 2019 Jun 19. J Mammary Gland Biol Neoplasia. 2019. PMID: 31218575
References
- Byrne C., Tainsky, M., and Fuchs, E. 1994. Programming gene expression in developing epidermis. Development 120: 2369–2383. - PubMed
- Cantor A.B. and Orkin, S.H. 2002. Transcriptional regulation of erythropoiesis: An affair involving multiple partners. Oncogene 21: 3368–3376. - PubMed
- Carlsson P., Waterman, M.L., and Jones, K.A. 1993. The hLEF/TCF-1 α HMG protein contains a context-dependent transcriptional activation domain that induces the TCR α enhancer in T cells. Genes & Dev. 7: 2418–2430. - PubMed
- Chan E.F., Gat, U., McNiff, J.M., and Fuchs, E. 1999. A common human skin tumour is caused by activating mutations in β-catenin. Nat. Genet. 21: 410–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM028896/GM/NIGMS NIH HHS/United States
- T32 GM007281/GM/NIGMS NIH HHS/United States
- 5T32 GM 07281/GM/NIGMS NIH HHS/United States
- GM 28896/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases