Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss - PubMed (original) (raw)
Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss
Beth A A Weaver et al. J Cell Biol. 2003.
Abstract
Centromere-associated protein-E (CENP-E) is an essential mitotic kinesin that is required for efficient, stable microtubule capture at kinetochores. It also directly binds to BubR1, a kinetochore-associated kinase implicated in the mitotic checkpoint, the major cell cycle control pathway in which unattached kinetochores prevent anaphase onset. Here, we show that single unattached kinetochores depleted of CENP-E cannot block entry into anaphase, resulting in aneuploidy in 25% of divisions in primary mouse fibroblasts in vitro and in 95% of regenerating hepatocytes in vivo. Without CENP-E, diminished levels of BubR1 are recruited to kinetochores and BubR1 kinase activity remains at basal levels. CENP-E binds to and directly stimulates the kinase activity of purified BubR1 in vitro. Thus, CENP-E is required for enhancing recruitment of its binding partner BubR1 to each unattached kinetochore and for stimulating BubR1 kinase activity, implicating it as an essential amplifier of a basal mitotic checkpoint signal.
Figures
Figure 1.
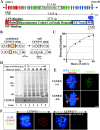
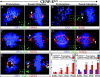
Murine CENP-E is efficiently removed after recombinase-mediated excision of the single murine CENP-E gene. (A) Schematic of the murine CENP-E gene, mRNA, and protein. The 46 exons of the CENP-E gene span 61.5 kb and encode a 7,425-nucleotide mRNA that produces a 2,474-aa protein. Apparent functional domains are labeled. KT, kinetochore; MT, microtubule; purple circles denote cdc2–cyclin B consensus phosphorylation sites. (Red) The kinesin-like motor region and associated exons; (orange) exon 4 encodes the ATP-binding consensus site of the motor domain and is selectively deleted (see B); (green) the discontinuous α-helical coiled-coil stalk region and associated exons; (blue) the carboxy-terminal globular domain and associated exons; CAAX, the terminal CAAX motif that may direct farnesylation. (B) Conversion of the conditional CENP-E allele to the null allele by Cre recombinase–mediated excision of exon 4 (exon P, shown in orange), due to lox P sites (denoted by triangles) incorporated into the adjacent introns, which introduces a premature stop codon at aa 82 (out of 2,474). (C) Real-time PCR analysis of a single experiment indicates that excision of the CENP-E gene reaches >90% by 48 h after addition of AdCre. (D) Immunoblot showing that CENP-E protein levels are diminished ∼16-fold in CENP-EΔ/Δ cells (lane 8) as compared with control cells (lane 3). Lane 1 is a 16-fold dilution of lane 3. Coomassie stain is shown as a loading control. (E) Immunofluorescence detection of CENP-E (green) in mitotic CENP-E+/+ or CENP-EΔ/Δ cells. CENP-EΔ/Δ MEFs acquire misaligned chromosomes that appear abnormally close to the spindle poles (white arrow). Tubulin, red; DNA stained with DAPI, blue. Bar, 2.5 μm. (F) Immunofluorescence detection of CENP-E (red) in mitotic CENP-E+/+ or CENP-EΔ/Δ cells. The CENP-E image in the CENP-EΔ/Δ cells was exposed 10 times longer than the CENP-E image in the CENP-E+/+ cells. CENP-E is still undetectable at kinetochores (marked by BubR1, green) in CENP-EΔ/Δ cells. DAPI, blue.
Figure 2.
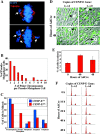
CENP-E Δ**/**Δ MEFs do not mount a robust cell cycle arrest despite the presence of unattached kinetochores. (A) Immunofluorescence of CENP-EΔ/Δ cells (48 h after addition of AdCre). Arrows denote polar chromosomes. DAPI, blue; tubulin, red. (B) Histogram showing the number of polar chromosomes per pseudo metaphase cell. (C) Graph showing the percentage of cells in various stages of mitosis. CENP-E+/+ cells are depicted by blue bars and CENP-EΔ/Δ cells are depicted by red bars. Pseudo metaphase cells are those in which the majority of chromosomes are aligned at the metaphase plate, but one or more chromosomes are at the poles. (D) Live cells whose DNA has been visualized by Hoechst 33258 do not exhibit marked mitotic arrest due to loss of CENP-E. Arrows denote mitotic cells. (E) Quantitation of the mitotic index of MEFs with one conditional and one null allele of CENP-E (CENP-EloxP/Δ) treated with AdCre for 0, 40, and 48 h. At least 1,000 MEFs were counted at each time point. (F) FACS® profiles of CENP-E+/+ cells (left column) and CENP-EloxP/Δ cells (right column) stained with propidium iodide at various times after addition of AdCre, indicating that loss of CENP-E does not induce robust mitotic arrest.
Figure 3.
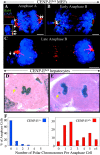
Absence of CENP-E causes cells to enter anaphase in the presence of one or a few polar chromosomes both in vitro and in vivo. (A–C) Immunofluorescence images of primary CENP-EΔ/Δ MEFs that have entered and proceeded through anaphase in the presence of one (B and C) or two (A) polar chromosomes that were never properly bioriented and aligned and will be missegregated. Bub1, green; DAPI, blue; tubulin, red. Bars, 2.5 μm. The inset in A is an enlargement of the polar chromosome at the left spindle pole showing the double dot pattern of Bub1 staining on the paired sister chromatids. (D and E) Regenerating hepatocytes that have entered (D) and proceeded through (E) anaphase in the presence of polar chromosomes (yellow arrows) in vivo after AdCre-mediated deletion of the single functional CENP-E gene in CENPElox P/Δ mice. (F) Histograms showing the number of polar chromosomes per anaphase figure in CENP-E+/+ (blue bars) and CENP-EΔ/Δ (red bars) hepatocytes in vivo. CENP-EΔ/Δ numbers were normalized to reflect the 70% excision rate.
Figure 4.
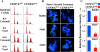
Microtubule depolymerization causes CENP-E Δ**/**Δ cells to arrest in mitosis with reduced amounts of BubR1, Mad1, and Mad2 on their kinetochores. (A) FACS® profile of propidium iodide–stained CENP-E+/+ cells (left column) and CENP-EΔ/Δ cells (right column) after microtubule depolymerization with colcemid or microtubule stabilization with taxol. 75% of these primary cells are cycling. (B) Immunofluorescence of CENP-E+/+ cells (left column) and CENP-EΔ/Δ cells (right column) after a 30-min treatment with 200 ng/ml colcemid, showing that CENP-E is required for maximal kinetochore targeting of BubR1, Mad1, and Mad2 (green). DNA is shown in blue. Bar, 2.5 μm. (C) Quantitation of the normalized integrated intensities of the kinetochore signals in B. ≥800 kinetochores from ≥10 cells were quantitated for each bar. Error bars represent standard errors. **, P < 0.001.
Figure 5.
CENP-E is required for efficient kinetochore targeting of BubR1, Mad1, and Mad2. (A–E′) Kinetochore localization of CENP-E (A and A′), BubR1 (B and B′), Mad1 (C and C′), Mad2 (D and D′), and Bub1 (E and E′) in CENP-E+/+ (A–E) and CENP-EΔ/Δ (A′–E′) cells. Kinetochore proteins are shown in green. DNA, blue; tubulin, red. Bar, 2.5 μm. (B′′–E′′) Quantitation of the normalized integrated intensity of BubR1 (B′′), Mad1 (C′′), Mad2 (D′′), and Bub1 (E′′) signals at kinetochores in CENP-E+/+ (blue bars) and CENP-EΔ/Δ (red bars) prometaphase and metaphase cells. Kinetochore signals on aligned chromosomes in CENP-EΔ/Δ pseudo metaphase cells are also shown. 20 –1,750 kinetochores from 2 to 19 different cells were quantitated for each bar. *, P < 0.05; **, P < 0.001. Error bars represent standard error.
Figure 6.
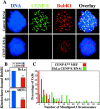
Kinetochores on polar chromosomes exhibit a specific defect in recruiting BubR1. (A–E) Polar kinetochores in pseudo metaphase cells (right) have higher levels of Bub1 (C), Mad1 (B and D), and Mad2 (E), but not BubR1 (A and B) than prometaphase kinetochores (left). Insets show higher magnification images of designated kinetochores. Arrows indicate polar chromosomes. DNA, blue; tubulin, red; checkpoint proteins, green, except in B, in which Mad1 is green and BubR1 is in red. (F and G) Comparison of the normalized integrated intensity of BubR1, Bub1, Mad1, and Mad2 signals at kinetochores. Prometaphase kinetochores in CENP-EΔ/Δ cells (striped bars in F) or in CENP-E+/+ cells (blue bars in G) are compared with kinetochores of polar chromosomes in CENP-EΔ/Δ pseudo metaphase cells (red bars). At least 53 kinetochores from ≥12 different cells were quantified for each bar. *, P < 0.05; **, P < 0.001. Error bars represent standard errors.
Figure 7.
HeLa cells depleted of CENP-E recruit reduced amounts of BubR1 to their kinetochores and have many misaligned chromosomes. (A) HeLa cells depleted of CENP-E (green) by RNAi (bottom) recruit less BubR1 (red) to their kinetochores than HeLa cells treated with control RNAi (top) after a 30-min treatment with 200 ng/ml colcemid. DNA, blue. (B) Quantitation of the normalized integrated intensity of BubR1 signals at kinetochores in HeLa cells (top) and primary MEFs (bottom). Control kinetochores (blue bars) recruit more BubR1 than CENP-E–depleted kinetochores (red bars). ≥800 kinetochores from ≥10 cells were quantitated for each bar. **, P < 0.001. Error bars represent standard error. (C) Histogram showing the number of misaligned chromosomes per pseudo metaphase cell in CENP-EΔ/Δ MEFs (red bars) and in HeLa cells depleted of CENP-E by RNAi (green bars).
Figure 8.
Mammalian BubR1 kinase activity is stimulated by CENP-E. (A) BubR1 was immunoprecipitated from CENP-E+/+ (lanes 1–3) and CENP-EΔ/Δ MEFs (lanes 4–6), and was assayed for kinase activity using histone H1 as a substrate (top). Equivalent levels of BubR1 were confirmed by immunoblot (bottom). Lane 1 and lane 4, beads alone control; lane 2 and lane 5, BubR1 was immunoprecipitated from randomly cycling cells; lane 3 and lane 6, BubR1 was immunoprecipitated from cells enriched in mitosis by a 16-h treatment with 50 ng/ml of the microtubule-depolymerizing agent colcemid. (B) Purified hCENP-E interacts with purified GST-hBubR1 bound to GSH beads (lane 2, top), but not to GST alone (lane 2, bottom). 25% of input was loaded in lane 1. (C) Coomassie stain of purified recombinant human BubR1, kinase-dead BubR1 (KD-BubR1), CENP-E, and Cdc20 that were expressed in insect cells using baculovirus. (D) In vitro kinase activity of purified hBubR1 by itself (lane 3 and lane 5), KD-hBubR1 by itself (lane 1), BubR1 + hCdc20 (lane 6), BubR1 + hCENP-E (lane 4 and lane 7), and BubR1 kinase-dead + hCENP-E (lane 2). Histone H1 was used as a substrate. (E) Coomassie stains of GSTHis-hBubR1, His-KD-hBubR1, and hCENP-E that were expressed singly or in combination in Hi5 cells and purified using GSH-Sepharose (left) or Ni-NTA agarose (right). Proteins in all lanes except 1 and 4 were purified in the presence of okadaic acid and 2 mM ATP. (F) Model of CENP-E as an activator of BubR1 in checkpoint signaling. In the presence of CENP-E (green, left) unattached kinetochores recruit large amounts of Bub1 (red), Mad1 (yellow), Mad2 (blue), and BubR1 (purple) whose kinase activity is stimulated by CENP-E (purple star). The unattached kinetochores assemble large quantities of Cdc20-APC/C inhibitors, which permit each kinetochore to delay anaphase onset until it has become attached. In the absence of CENP-E (right), unattached kinetochores recruit reduced amounts of Mad1 (yellow), Mad2 (blue), and BubR1 (purple) with reduced kinase activity (no star). Each kinetochore produces fewer molecules of Cdc20-APC/C inhibitors, which are no longer sufficient to meet the threshold required to prevent premature anaphase onset.
Similar articles
- Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling.
Mao Y, Desai A, Cleveland DW. Mao Y, et al. J Cell Biol. 2005 Sep 12;170(6):873-80. doi: 10.1083/jcb.200505040. Epub 2005 Sep 6. J Cell Biol. 2005. PMID: 16144904 Free PMC article. - Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores.
Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Ditchfield C, et al. J Cell Biol. 2003 Apr 28;161(2):267-80. doi: 10.1083/jcb.200208091. J Cell Biol. 2003. PMID: 12719470 Free PMC article. - A conserved CENP-E region mediates BubR1-independent recruitment to the outer corona at mitotic onset.
Weber J, Legal T, Lezcano AP, Gluszek-Kustusz A, Paterson C, Eibes S, Barisic M, Davies OR, Welburn JPI. Weber J, et al. Curr Biol. 2024 Mar 11;34(5):1133-1141.e4. doi: 10.1016/j.cub.2024.01.042. Epub 2024 Feb 13. Curr Biol. 2024. PMID: 38354735 - Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling.
Cleveland DW, Mao Y, Sullivan KF. Cleveland DW, et al. Cell. 2003 Feb 21;112(4):407-21. doi: 10.1016/s0092-8674(03)00115-6. Cell. 2003. PMID: 12600307 Review. - A Condensed View of the Chromosome Passenger Complex.
Trivedi P, Stukenberg PT. Trivedi P, et al. Trends Cell Biol. 2020 Sep;30(9):676-687. doi: 10.1016/j.tcb.2020.06.005. Epub 2020 Jul 17. Trends Cell Biol. 2020. PMID: 32684321 Free PMC article. Review.
Cited by
- GPER1 links estrogens to centrosome amplification and chromosomal instability in human colon cells.
Bühler M, Fahrländer J, Sauter A, Becker M, Wistorf E, Steinfath M, Stolz A. Bühler M, et al. Life Sci Alliance. 2022 Nov 16;6(1):e202201499. doi: 10.26508/lsa.202201499. Print 2023 Jan. Life Sci Alliance. 2022. PMID: 36384894 Free PMC article. - Evolutionarily conserved protein ERH controls CENP-E mRNA splicing and is required for the survival of KRAS mutant cancer cells.
Weng MT, Lee JH, Wei SC, Li Q, Shahamatdar S, Hsu D, Schetter AJ, Swatkoski S, Mannan P, Garfield S, Gucek M, Kim MK, Annunziata CM, Creighton CJ, Emanuele MJ, Harris CC, Sheu JC, Giaccone G, Luo J. Weng MT, et al. Proc Natl Acad Sci U S A. 2012 Dec 26;109(52):E3659-67. doi: 10.1073/pnas.1207673110. Epub 2012 Dec 10. Proc Natl Acad Sci U S A. 2012. PMID: 23236152 Free PMC article. - A mitotic septin scaffold required for Mammalian chromosome congression and segregation.
Spiliotis ET, Kinoshita M, Nelson WJ. Spiliotis ET, et al. Science. 2005 Mar 18;307(5716):1781-5. doi: 10.1126/science.1106823. Science. 2005. PMID: 15774761 Free PMC article. - Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F.
Bomont P, Maddox P, Shah JV, Desai AB, Cleveland DW. Bomont P, et al. EMBO J. 2005 Nov 16;24(22):3927-39. doi: 10.1038/sj.emboj.7600848. Epub 2005 Oct 27. EMBO J. 2005. PMID: 16252009 Free PMC article. - Cenp-F (mitosin) is more than a mitotic marker.
Varis A, Salmela AL, Kallio MJ. Varis A, et al. Chromosoma. 2006 Aug;115(4):288-95. doi: 10.1007/s00412-005-0046-0. Epub 2006 Mar 25. Chromosoma. 2006. PMID: 16565862 Review.
References
- Abrieu, A., J.A. Kahana, K.W. Wood, and D.W. Cleveland. 2000. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 102:817–826. - PubMed
- Ashar, H.R., L. James, K. Gray, D. Carr, S. Black, L. Armstrong, W.R. Bishop, and P. Kirschmeier. 2000. Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J. Biol. Chem. 275:30451–30457. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases