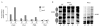Gcn4 occupancy of open reading frame regions results in the recruitment of chromatin-modifying complexes but not the mediator complex - PubMed (original) (raw)
Gcn4 occupancy of open reading frame regions results in the recruitment of chromatin-modifying complexes but not the mediator complex
Irini Topalidou et al. EMBO Rep. 2003 Sep.
Abstract
Eukaryotic transcriptional activators usually recognize short DNA motifs, which are not only located within promoter regions, but also scattered throughout the genome. Assuming that the function of activators at non-promoter regions is wasteful and perhaps harmful, one can ask whether such binding is somehow prevented or if transcription is blocked at a downstream step. Here, we show that the yeast transcriptional activator Gcn4 is associated in vivo with several non-promoter euchromatic sites. This association results in the recruitment of the SAGA (Spt3/Ada/Gcn5/acetyltransferase) complex and the consequent activity of the Gcn5 histone acetyltransferase. The functional recruitment of the Swi/Snf nucleosome-remodelling complex was also evident at sites located in positioned nucleosomes. We show that this assemblage of coactivator complexes is not productive because of the absence of core promoter elements, other than the TATA box, that are required for stable mediator recruitment.
Figures
Figure 1
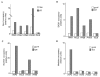
Recruitment of the Gcn4 transcriptional activator and coactivators at non-promoter chromosomal regions. (A) In vivo occupancy by Gcn4 of the TRP3 promoter and target sites located at the indicated regions and after growth in minimal (Min) or histidine starvation (AT) media. Levels shown on the y axis reflect the enrichment of the DNA relative to a region of the PHO5 open reading frame after correction for the ratios of amplification achieved using input DNA. (B) Occupancy of the same sites by the SAGA (Spt3/Ada/Gcn5/acetyltransferase) complex, monitored using a MYC-tagged Spt3 component, in relation to the presence (WT) or absence (gcn4) of Gcn4. (C) Occupancy of Swi/Snf in relation to the presence or absence of Gcn4, probed through a MYC-tagged Snf2 component. (D) Occupancy of the mediator complex in relation to the presence or absence of Gcn4, probed using a MYC-tagged Srb4 component. In the experiments presented in (B–D), strains were grown in conditions of histidine starvation.
Figure 2
The high level of occupancy of the PHO8 site by Gcn4 does not contribute to the regulation of PHO8 expression, or that of the upstream_KRE2_ gene. (A) The messenger RNA levels of the PHO8 gene from cells grown in either high- or low-phosphate media (HP and LP, respectively) are not affected by the presence of either low (WT/M) or high (WT/AT) levels of Gcn4, or by the absence (gcn4) of this protein. (B) mRNA levels of the KRE2 gene in wild-type (WT) and_gcn4_ strains grown in either minimal (M) or histidine starvation (AT) media. Actin mRNA (ACT1) was used as loading control in both experiments.
Figure 3
Functional consequences of coactivator recruitment. (A) Gcn5-dependent acetylation of nucleosomal histones located at the regions indicated was measured at low (M) or high (AT) Gcn4 concentrations by chromatin immunoprecipitation using antibodies raised against a diacetylated form of histone H3. Numbers indicate the enrichment of the DNA relative to a telomeric region (TEL) and corrected for the level of amplification achieved using input DNA. TEL (Vogelauer et al., 2000) was used as a control as it is heterochromatic and therefore under-acetylated. (B) The diagram in the left panel shows the mapped positions of nucleosomes around the Gcn4-response element (GCRE) site of the PHO8 gene. Gcn4-dependent remodelling of the nucleosomes positioned on either side of the GCRE located in the PHO8 open reading frame is shown in the left panel. Remodelling was monitored in wild-type (WT) or snf2 strains by determining the sensitivity to micrococcal nuclease and indirect end-labelling. M and AT indicate cells with low and high amounts of Gcn4, respectively. N is the micrococcal-nuclease cleavage pattern of naked DNA. By contrast, similar experiments for the POL2 site (right panel) showed the absence of positioned nucleosomes, as indicated by the identical patterns of micrococcal nuclease cleavages in nuclear (M and AT) or naked (N) DNA.
Figure 4
An engineered non-promoter site defines the requirements for coactivator recruitment. (A) The top left panel shows a representation of the PHO5 promoter with a Gcn4-response element (GCRE) site introduced into the linker between nucleosomes −2 and −1. Also indicated are the _Cla_I cleavage site used for monitoring nucleosome remodelling, the TATA box and the transcription initiation site (arrow). The right panel shows that in cells growing under histidine limitation (high levels of Gcn4), the wild-type promoter is occupied by Gcn4, the mediator complex (Srb4), SAGA (Spt3) and TATA-binding protein (TBP). The bottom left panel shows that Snf2 is required for remodelling of nucleosome −2. Remodelling was assayed by determining the accessibility to _Cla_I of cells containing low (M) or high (AT) amounts of Gcn4 in wild-type (WT) or snf2 strains. P indicates the protected DNA; U indicates one fragment that results from cleavage by the enzyme used and secondary digestions. (B) Mutating the TATA element (top left panel) affects only the recruitment of TBP. (C) The absence of all promoter elements and downstream open reading frame sequences (top left panel) allows the recruitment of SAGA.
Similar articles
- Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo.
Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG. Govind CK, et al. Mol Cell Biol. 2005 Jul;25(13):5626-38. doi: 10.1128/MCB.25.13.5626-5638.2005. Mol Cell Biol. 2005. PMID: 15964818 Free PMC article. - Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes.
Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Hassan AH, et al. Cell. 2002 Nov 1;111(3):369-79. doi: 10.1016/s0092-8674(02)01005-x. Cell. 2002. PMID: 12419247 - Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex.
Biswas D, Imbalzano AN, Eriksson P, Yu Y, Stillman DJ. Biswas D, et al. Mol Cell Biol. 2004 Sep;24(18):8312-21. doi: 10.1128/MCB.24.18.8312-8321.2004. Mol Cell Biol. 2004. PMID: 15340090 Free PMC article. - Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.
Wallberg AE, Flinn EM, Gustafsson JA, Wright AP. Wallberg AE, et al. Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review. - Remodeling chromatin structures for transcription: what happens to the histones?
Steger DJ, Workman JL. Steger DJ, et al. Bioessays. 1996 Nov;18(11):875-84. doi: 10.1002/bies.950181106. Bioessays. 1996. PMID: 8939065 Review.
Cited by
- A role for gcn5-mediated global histone acetylation in transcriptional regulation.
Imoberdorf RM, Topalidou I, Strubin M. Imoberdorf RM, et al. Mol Cell Biol. 2006 Mar;26(5):1610-6. doi: 10.1128/MCB.26.5.1610-1616.2006. Mol Cell Biol. 2006. PMID: 16478983 Free PMC article. - Revisiting the model for coactivator recruitment: Med15 can select its target sites independent of promoter-bound transcription factors.
Mindel V, Brodsky S, Yung H, Manadre W, Barkai N. Mindel V, et al. Nucleic Acids Res. 2024 Nov 11;52(20):12093-12111. doi: 10.1093/nar/gkae718. Nucleic Acids Res. 2024. PMID: 39187372 Free PMC article. - Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p.
Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG. Qiu H, et al. Mol Cell Biol. 2005 May;25(9):3461-74. doi: 10.1128/MCB.25.9.3461-3474.2005. Mol Cell Biol. 2005. PMID: 15831453 Free PMC article. - Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment.
Topalidou I, Papamichos-Chronakis M, Thireos G, Tzamarias D. Topalidou I, et al. EMBO J. 2004 May 5;23(9):1943-8. doi: 10.1038/sj.emboj.7600199. Epub 2004 Apr 1. EMBO J. 2004. PMID: 15057269 Free PMC article. - Mediator influences telomeric silencing and cellular life span.
Zhu X, Liu B, Carlsten JO, Beve J, Nyström T, Myers LC, Gustafsson CM. Zhu X, et al. Mol Cell Biol. 2011 Jun;31(12):2413-21. doi: 10.1128/MCB.05242-11. Epub 2011 Apr 11. Mol Cell Biol. 2011. PMID: 21482672 Free PMC article.
References
- Brown C.E., Howe L., Sousa K., Alley S.C., Carrozza M.J., Tan S. & Workman J.L. ( 2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science, 292, 2333–2337. - PubMed
- Bryant G.O. & Ptashne M. ( 2003) Independent recruitment in vivo by gal4 of two complexes required for transcription. Mol. Cell, 11, 1301–1309. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials