The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies - PubMed (original) (raw)
The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies
Yutong Xue et al. Proc Natl Acad Sci U S A. 2003.
Abstract
ATRX syndrome is characterized by X-linked mental retardation associated with alpha-thalassemia. The gene mutated in this disease, ATRX, encodes a plant homeodomain-like finger and a SWI2/SNF2-like ATPase motif, both of which are often found in chromatin-remodeling enzymes, but ATRX has not been characterized biochemically. By immunoprecipitation from HeLa extract, we found that ATRX is in a complex with transcription cofactor Daxx. The following evidence supports that ATRX and Daxx are components of an ATP-dependent chromatin-remodeling complex: (i) Daxx and ATRX can be coimmunoisolated by antibodies specific for each protein; (ii) a proportion of Daxx cofractionates with ATRX as a complex of 1 MDa by gel-filtration analysis; (iii) in extract from cells of a patient with ATRX syndrome, the level of the Daxx-ATRX complex is correspondingly reduced; (iv) a proportion of ATRX and Daxx colocalize in promyelocytic leukemia nuclear bodies, with which Daxx had previously been located; and (v) the ATRX complex displays ATP-dependent activities that resemble those of other chromatin-remodeling complexes, including triple-helix DNA displacement and alteration of mononucleosome disruption patterns. But unlike the previously described SWI/SNF or NURD complexes, the ATRX complex does not randomize DNA phasing of the mononucleosomes, suggesting that it may remodel chromatin differently. Taken together, the results suggest that ATRX functions in conjunction with Daxx in a novel chromatin-remodeling complex. The defects in ATRX syndrome may result from inappropriate expression of genes controlled by this complex.
Figures
Fig. 4.
The ATRX complex has an ATP-dependent triple-helix displacement activity. (a and b) Autoradiographs showing that the ATRX–Daxx complex displaces a triple helix in the presence of ATP. The positions of the labeled triple-helix substrate and the displaced third strand in the gel are depicted at the left. The complexes isolated by different Abs to ATRX, Daxx, and human SWI/SNF are shown at the top. Polypeptides isolated by mock immunoprecipitation by using either protein A beads (PnA) or preimmune serum (mock IP) are also indicated. Human SWI/SNF in our hands does not show significant activity in this assay. To serve as a positive control, recombinant STH1 (a generous gift of B. Cairns, Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City) was used. (c) Autoradiograph showing that the ATRX–Daxx complex does not displace a blunt triple helix. The substrates used are indicated at the bottom. The substrates used in the blunt triplex assay contain two bands. The major band (with the faster mobility) represents the blunt triplex. The minor band (with slower mobility) represents a triplex substrate with one blunt end and one overhang end of 17-bp double-stranded DNA. This overhang was designed to help annealing of the double-stranded DNA and was supposed to be removed by restriction digestion, but it was not completely removed in the current experiment. (d) Autoradiograph showing the results of a double-helix displacement assay. The duplex substrate and the displaced oligo are illustrated at the left. The BLM (Bloom Syndrome gene product) DNA helicase complex was used as a positive control (18). (e) Immunoblot showing that Daxx was completely removed by washing with buffer containing 0.5 or 0.75 M salt, as shown at the top. (f) Autoradiograph showing that dissociation of Daxx does not affect triplex unwinding activity of ATRX.
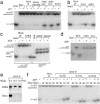
Fig. 1.
Purification of an ATRX-associated complex from HeLa nuclear extract. (a) Silver-stained SDS gels showing polypeptides immunoisolated by three different ATRX Abs (NP5, D19, and C16) from nuclear extract. Mock immunoprecipitation was done by using either protein A beads alone (lane 2) or a preimmune (PI) serum (lane 5). MS has been used for identification of ATRX and its associated polypeptides in all three preparations. The number of peptides that matched the indicated protein and the percentage of these peptides among total peptides obtained from matrix-assisted laser desorption ionization–time-of-flight analysis are shown in parentheses for ATRX and Daxx. The other polypeptides (marked by lines) appeared to either loosely associate with ATRX or associate with ATRX through DNA by subsequent analysis. (b) Immunoblot confirming the presence of Daxx in the polypeptides immunoisolated by ATRX Ab. NE, nuclear extract; SN, supernatant; IP, immunoprecipitate. The lower Daxx band in nuclear extract could represent a posttranslationally modified version of Daxx that associates poorly with SDS. (c) Silver-stained gel showing polypeptides immunoisolated by an ATRX Ab from the ATRX peak fractions after Superose 6 fractionation of nuclear extract (Fig. 2_c_). The presence or absence of ethidium bromide (EtBr) is indicated. Note that p90 and p70 polypeptides are lost in IP in the presence of EtBr, hinting that they may associate with ATRX through DNA. Also, Daxx becomes substoichiometric compared to ATRX, which could be caused by partial dissociation of the ATRX–Daxx complex during Superose 6 fractionation. (d) Immunoblot showing that association between ATRX and Daxx is not through DNA.
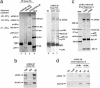
Fig. 2.
Daxx and ATRX form a complex, and the level of this complex is significantly decreased in a ATRX patient cell line. (a) Silver-stained SDS gel showing the polypeptides immunoisolated by a Daxx Ab compared to those by the ATRX Ab. All polypeptides marked by an asterisk have been identified by MS. The number of peptides that match the indicated protein and the percentage of these peptides among total peptides obtained from matrix-assisted laser desorption ionization–time-of-flight analysis are shown in parentheses for ATRX and Daxx. (b) Immunoblot confirming the presence of ATRX in the polypeptides isolated by the Daxx Ab. (c) Immunoblot showing the Superose 6 gel-filtration profile of ATRX and Daxx in nuclear extracts prepared from either HeLa cells (Upper) or a cell line derived from an ATRX patient (Lower). The peaks corresponding to proteins with known molecular masses are denoted at the bottom. Note that the peak of the ATRX–Daxx complex (fraction 20, indicated by an arrowhead) is significantly reduced in the ATRX patient cell line. (d) Immunoblot analysis showing the levels of ATRX (Upper) and Daxx (Lower) in the ATRX patient cell line in comparison to those in HeLa cells.
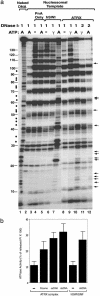
Fig. 3.
The ATRX complex alters the DNase I digestion pattern of a nucleosome in the presence of ATP. (a) Autoradiograph showing the results of the mononucleosome disruption assay. Complexes isolated by protein A alone (PnA) or Abs against ATRX and human SWI/SNF (hSWI) are indicated at the top. The templates used and the presence of ATP (A) or ATP-γ-S (γ) are indicated. The amounts of DNase used, 1 and 2, represent 0.2 and 0.4 units, respectively. The solid arrows mark the nucleotides whose digestion was enhanced by the ATRX–Daxx complex in the presence of ATP. The solid dots denote the nucleotides between the 10-bp ladders whose digestion was stimulated by human SWI/SNF but not ATRX–Daxx. The solid lines mark the positions of the nucleotides in 5S DNA. The 10-bp ladders can be observed between nucleotides 10 and 130 of 5S DNA. (b) Graphic presentation showing that the ATRX complex has DNA- or nucleosome-stimulated ATPase activity (shown in percentage of released inorganic phosphate from total ATP multiplied by 100). The ATPase activity of mock immunoprecipitation (using preimmune serum) is indistinguishable from that of the background (using protein A beads), which was subtracted during calculation. The presence of nucleosomes (N′some), single-stranded DNA (ssDNA), and double-stranded DNA (dsDNA) is indicated.
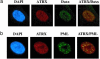
Fig. 5.
ATRX and Daxx colocalize in PML nuclear bodies. (a) Immunofluorescence images showing colocalization of ATRX and Daxx in human fibroblast cell lines. Images for each protein and the merged image are shown. (b) Immunofluorescence images showing colocalization of ATRX with PML in human fibroblasts. The nucleus was costained with 4′,6-diamidino-2-phenylindole (DAPI).
Similar articles
- A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein.
Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X. Tang J, et al. J Biol Chem. 2004 May 7;279(19):20369-77. doi: 10.1074/jbc.M401321200. Epub 2004 Feb 27. J Biol Chem. 2004. PMID: 14990586 - Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX.
Ishov AM, Vladimirova OV, Maul GG. Ishov AM, et al. J Cell Sci. 2004 Aug 1;117(Pt 17):3807-20. doi: 10.1242/jcs.01230. Epub 2004 Jul 13. J Cell Sci. 2004. PMID: 15252119 - The Daxx/Atrx Complex Protects Tandem Repetitive Elements during DNA Hypomethylation by Promoting H3K9 Trimethylation.
He Q, Kim H, Huang R, Lu W, Tang M, Shi F, Yang D, Zhang X, Huang J, Liu D, Songyang Z. He Q, et al. Cell Stem Cell. 2015 Sep 3;17(3):273-86. doi: 10.1016/j.stem.2015.07.022. Cell Stem Cell. 2015. PMID: 26340527 Free PMC article. - ATRX and DAXX: Mechanisms and Mutations.
Dyer MA, Qadeer ZA, Valle-Garcia D, Bernstein E. Dyer MA, et al. Cold Spring Harb Perspect Med. 2017 Mar 1;7(3):a026567. doi: 10.1101/cshperspect.a026567. Cold Spring Harb Perspect Med. 2017. PMID: 28062559 Free PMC article. Review. - Emerging roles of ATRX in cancer.
Watson LA, Goldberg H, Bérubé NG. Watson LA, et al. Epigenomics. 2015;7(8):1365-78. doi: 10.2217/epi.15.82. Epub 2015 Dec 8. Epigenomics. 2015. PMID: 26646632 Review.
Cited by
- The Multiple Facets of ATRX Protein.
Valenzuela M, Amato R, Sgura A, Antoccia A, Berardinelli F. Valenzuela M, et al. Cancers (Basel). 2021 May 5;13(9):2211. doi: 10.3390/cancers13092211. Cancers (Basel). 2021. PMID: 34062956 Free PMC article. Review. - Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity.
Jurak I, Silverstein LB, Sharma M, Coen DM. Jurak I, et al. J Virol. 2012 Sep;86(18):10093-102. doi: 10.1128/JVI.00930-12. Epub 2012 Jul 11. J Virol. 2012. PMID: 22787211 Free PMC article. - Protein complex of Drosophila ATRX/XNP and HP1a is required for the formation of pericentric beta-heterochromatin in vivo.
Emelyanov AV, Konev AY, Vershilova E, Fyodorov DV. Emelyanov AV, et al. J Biol Chem. 2010 May 14;285(20):15027-15037. doi: 10.1074/jbc.M109.064790. Epub 2010 Feb 13. J Biol Chem. 2010. PMID: 20154359 Free PMC article. - A Novel Mutation in ATRX Causes Alpha-Thalassemia X-Linked Intellectual Disability Syndrome in a Han Chinese Family.
Wu S, Zheng Y, Xu C, Fu J, Xiong F, Yang F. Wu S, et al. Front Pediatr. 2022 Jan 20;9:811812. doi: 10.3389/fped.2021.811812. eCollection 2021. Front Pediatr. 2022. PMID: 35127601 Free PMC article. - Reciprocal regulation of Daxx and PIK3CA promotes colorectal cancer cell growth.
Huang YS, Wu CC, Chang CC, Huang SF, Kuo HY, Shih HM. Huang YS, et al. Cell Mol Life Sci. 2022 Jun 19;79(7):367. doi: 10.1007/s00018-022-04399-8. Cell Mol Life Sci. 2022. PMID: 35718818 Free PMC article.
References
- Krebs, J. E. & Peterson, C. L. (2000) Crit. Rev. Eukaryotic Gene Expression 10, 1–12. - PubMed
- Neely, K. & Workman, J. (2002) Biochim. Biophys. Acta 1603, 19–29. - PubMed
- Kwon, H., Imbalzano, A. N., Khavari, P. A., Kingston, R. E. & Green, M. R. (1994) Nature 370, 477–481. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases