The Drosophila melanogaster BTB proteins bric à brac bind DNA through a composite DNA binding domain containing a pipsqueak and an AT-Hook motif - PubMed (original) (raw)
The Drosophila melanogaster BTB proteins bric à brac bind DNA through a composite DNA binding domain containing a pipsqueak and an AT-Hook motif
Corinne Lours et al. Nucleic Acids Res. 2003.
Abstract
The bric à brac (bab) locus is composed of two paralogous genes, bab1 and bab2, in Drosophila melanogaster. Bab1 and Bab2 are nuclear proteins that contain a broad complex, tramtrack, bric à brac/poxviruses and zinc-finger (BTB/POZ) domain. Many BTB/POZ proteins are transcriptional regulators of which the majority contain C(2)H(2) zinc-finger motifs. There is no detectable zinc-finger motif in either Bab protein. However, they share the Bab conserved domain (BabCD) that is highly conserved between Bab1 and Bab2, and the Bab proteins of several other species, e.g. Anopheles gambiae, Apis mellifera and Drosophila virilis. Here we show that Bab2 binds to several discrete sites on polytene chromosomes including the bab locus, and that the BabCD of both Bab1 and Bab2 binds in vitro to the cis-regulatory regions of bab1 and bab2. Our results indicate that the BabCD binds to A/T-rich regions and that its optimum binding sites contain TA or TAA repeats. The BabCD is a composite DNA binding domain with a psq motif and an AT-Hook motif; both motifs are required for DNA binding activity. Structural similarities suggest that the BabCD may bind to DNA in a similar manner as some prokaryotic recombinases.
Figures
Figure 1
Bab2 proteins bind to several loci on polytene chromosomes. (A) The Bab2 protein was detected on squashed salivary gland polytene chromosomes and the strongest binding sites were cytologically mapped. The arrow on the left arm of the third chromosome points to the bab locus at 61F1-2. Indicated on the right are the map positions of the 12 strongest binding sites that include a double band at the 24B/C boundary. (B) and (C) Tip of the left arm of the third chromosome stained with the anti-Bab2 antibody (B) or with a digoxigenin labeled bab2 cDNA (C).
Figure 2
The BabCD specifically binds to DNA in vitro. (A) GST–BabCD1 fusion protein and GST were produced in E.coli, purified and analyzed by SDS–PAGE electrophoresis, and stained with Coomassie blue. Total protein extracts were analyzed after induction (lanes 1 and 4) or without induction (lanes 2 and 5), and after purification (lanes 3 and 6). The molecular mass of the GST–BabCD1 fusion protein and GST are indicated in kDa on the right. (B) An 8 kb bab1 subclone was digested with HaeIII and the digestion products were incubated with either GST or GST–BabCD1 fusion protein bound to glutathione–agarose beads. The HaeIII digestion products (lane 1) and the fragments retained by GST (lane 2) or GST–BabCD1 (lane 3) were analyzed on 5% polyacrylamide gel. (C) A 2.5 kb bab2 subclone was digested with HaeIII and the digestion products were incubated with either GST, GST–BabCD1 or GST–BabCD2 fusion proteins bound to glutathione–agarose beads. The experiment is as in (B). Lane 1 shows the HaeIII digestion products, lanes 3 and 4 the fragments retained by the GST–BabCD1 and GST–BabCD2 proteins, respectively, and the control experiment with GST is shown in lane 2. The length of the DNA fragments retained by both the GST–BabCD1 and GST–BabCD2 protein are indicated in bp on the right.
Figure 3
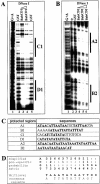
The BabCD binds to TA/TAA rich sites. A DNase I footprinting analysis was done with the 209 bp bab1 fragment (A) or the 220 bp bab2 fragment (B) as probes and the GST or GST–BabCD1 fusion proteins. ‘0’ indicates that the experiment was done without protein (Fig. 4B, lane 2). Two or three increasing dilutions of DNase I enzyme were used as indicated by the triangle above the lanes. Products were analyzed on a 7% polyacrylamide gel in parallel with a G + A sequence reaction (lane 1). (C) Sequences of the protected regions on the 209 bp bab1 fragment (C1 and D1), on a 347 bp bab1 fragment (A1 and B1, not shown) and on the 220 bp bab2 fragment (A2 and B2). The T and A bases are in bold. (D) CAST: oligonucleotides retained by the BabCD1 fusion protein were selected by IVPO. DNA from 27 plasmids were sequenced and the internal variable sequences were aligned using MEME version 3.0. The position-specific probability matrix and the multilevel consensus sequence calculated by MEME are shown.
Figure 4
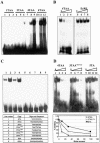
GST–BabCD fusion proteins bind to repeats of TA or TAA. (A) EMSAs with GST (lanes 1, 4, 7 and 10) or GST–BabCD1 (lanes 2, 3, 5, 6, 8, 9, 11 and 12) purified proteins and oligonucleotides containing one (lanes 1–3), two (lanes 4–6), three (lanes 7–9) or four (lanes 10–12) repeats of TAA. Lanes 3, 6, 9 and 12 contain 5-fold more protein than lanes 2, 5, 8 and 11. (B) GST–BabCD1 protein was incubated with radiolabeled 4TAA oligonucleotide, without (lanes 1 and 5) or with (lanes 2–4) increasing amounts of cold 4TAA oligonucleotide or EcRE binding site (lanes 6–8). Each competitor was added in an molar excess of 10-, 30- and 100-fold. (C) EMSAs with GST–BabCD1 proteins and different oligonucleotides in each lane. The name and the sequence of the corresponding oligonucleotides are listed in the table. (D) GST–BabCD1 protein was incubated with radiolabeled 4TAA double-stranded oligonucleotide without (lanes 1, 5 and 9) or with increasing amounts of cold 4TAA (lanes 2–4), 3TAA*TTA (lanes 6–8) or 5TA (lanes 10–12) oligonucleotides. The amounts of competitors used are 10, 30 and 100 molar excess. A graphical representation of the percentage of bound radiolabeled oligonucleotide 4TAA, quantitated by PhosphorImager analysis, is shown below. The amount of probe bound in the absence of a competitor was given the value 100%.
Figure 5
Structure and DNA binding activity of full-length or truncated versions of the BabCD. Designations of the truncated derivatives indicate the number of amino acid residues. The positions of the deletions in the BabCD1 amino acid sequence are indicated at the bottom. The psq and AT-Hook motif are indicated in each protein by black boxes. The DNA binding activity of each protein, determined by IVPO analysis of the 2.5 kb bab2 subclone that was digested with HaeIII, is indicated on the right.
Figure 6
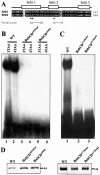
The AT-Hook and the psq motif are required for the DNA binding activity of the BabCD. (A) Indicated above the sequence alignment of the psq motif of Bab1 and Bab2 are the positions of three predicted alpha helices. The mutations made in helix 1 or in helix 2 of the psq domain of BabCD1 are shown below the sequences. (B) EMSAs with the 3TAA (lanes 1, 3 and 5) or the 4TAA (lanes 2, 4 and 6) oligonucleotides and the wild-type BabCD1 (lanes 1 and 2), or two psq domain mutants: BabCDAI576GP (lanes 2 and 4) and BabCDA590P (lanes 5 and 6). (C) EMSAs with the 4TAA oligonucleotide and the wild-type BabCD1 (lane 1), the BabCDAT-Hook1 (lane 2) or the BabCDAT-Hook2 (lane 3) mutants. (D) Coomassie blue-stained SDS–PAGE containing the indicated purified wild-type or mutant BabCD1 proteins. Their molecular mass is indicated in kDa on the right.
Similar articles
- The bric à brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila.
Couderc JL, Godt D, Zollman S, Chen J, Li M, Tiong S, Cramton SE, Sahut-Barnola I, Laski FA. Couderc JL, et al. Development. 2002 May;129(10):2419-33. doi: 10.1242/dev.129.10.2419. Development. 2002. PMID: 11973274 - The BTB/POZ domain of the regulatory proteins Bric à brac 1 (BAB1) and Bric à brac 2 (BAB2) interacts with the novel Drosophila TAF(II) factor BIP2/dTAF(II)155.
Pointud JC, Larsson J, Dastugue B, Couderc JL. Pointud JC, et al. Dev Biol. 2001 Sep 15;237(2):368-80. doi: 10.1006/dbio.2001.0358. Dev Biol. 2001. PMID: 11543621 - The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain.
Lehmann M, Siegmund T, Lintermann KG, Korge G. Lehmann M, et al. J Biol Chem. 1998 Oct 23;273(43):28504-9. doi: 10.1074/jbc.273.43.28504. J Biol Chem. 1998. PMID: 9774480 - Broad-complex, tramtrack, and bric-à-brac (BTB) proteins: Critical regulators of development.
Chaharbakhshi E, Jemc JC. Chaharbakhshi E, et al. Genesis. 2016 Oct;54(10):505-518. doi: 10.1002/dvg.22964. Epub 2016 Aug 22. Genesis. 2016. PMID: 27521773 Review. - The genetics and molecular biology of zeste in Drosophila melanogaster.
Pirrotta V. Pirrotta V. Adv Genet. 1991;29:301-48. doi: 10.1016/s0065-2660(08)60110-8. Adv Genet. 1991. PMID: 1763708 Review. No abstract available.
Cited by
- Cotton leaf curl Multan virus differentially regulates innate antiviral immunity of whitefly (Bemisia tabaci) vector to promote cryptic species-dependent virus acquisition.
Farooq T, Lin Q, She X, Chen T, Li Z, Yu L, Lan G, Tang Y, He Z. Farooq T, et al. Front Plant Sci. 2022 Nov 14;13:1040547. doi: 10.3389/fpls.2022.1040547. eCollection 2022. Front Plant Sci. 2022. PMID: 36452094 Free PMC article. - Transcription factors, chromatin proteins and the diversification of Hemiptera.
Vidal NM, Grazziotin AL, Iyer LM, Aravind L, Venancio TM. Vidal NM, et al. Insect Biochem Mol Biol. 2016 Feb;69:1-13. doi: 10.1016/j.ibmb.2015.07.001. Epub 2015 Jul 29. Insect Biochem Mol Biol. 2016. PMID: 26226651 Free PMC article. - Contrasting patterns of sequence evolution at the functionally redundant bric à brac paralogs in Drosophila melanogaster.
Bickel RD, Schackwitz WS, Pennacchio LA, Nuzhdin SV, Kopp A. Bickel RD, et al. J Mol Evol. 2009 Aug;69(2):194-202. doi: 10.1007/s00239-009-9265-y. Epub 2009 Jul 29. J Mol Evol. 2009. PMID: 19639236 Free PMC article. - Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome.
Vogelmann J, Le Gall A, Dejardin S, Allemand F, Gamot A, Labesse G, Cuvier O, Nègre N, Cohen-Gonsaud M, Margeat E, Nöllmann M. Vogelmann J, et al. PLoS Genet. 2014 Aug 28;10(8):e1004544. doi: 10.1371/journal.pgen.1004544. eCollection 2014 Aug. PLoS Genet. 2014. PMID: 25165871 Free PMC article. - The Bric-à-Brac BTB/POZ transcription factors are necessary in niche cells for germline stem cells establishment and homeostasis through control of BMP/DPP signaling in the Drosophila melanogaster ovary.
Miscopein Saler L, Hauser V, Bartoletti M, Mallart C, Malartre M, Lebrun L, Pret AM, Théodore L, Chalvet F, Netter S. Miscopein Saler L, et al. PLoS Genet. 2020 Nov 5;16(11):e1009128. doi: 10.1371/journal.pgen.1009128. eCollection 2020 Nov. PLoS Genet. 2020. PMID: 33151937 Free PMC article.
References
- Bardwell V.J. and Treisman,R. (1994) The POZ domain: a conserved protein–protein interaction motif. Genes Dev., 8, 1664–1677. - PubMed
- Kobayashi A., Yamagiwa,H., Hoshino,H., Muto,A., Sato,K., Morita,M., Hayashi,N., Yamamoto,M. and Igarashi,K. (2000) A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol., 20, 1733–1746. - PMC - PubMed
- Tsukiyama T., Becker,P.B. and Wu,C. (1994) ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature, 367, 525–532. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases