Sex genes for genomic analysis in human brain: internal controls for comparison of probe level data extraction - PubMed (original) (raw)
Comparative Study
Sex genes for genomic analysis in human brain: internal controls for comparison of probe level data extraction
Hanga C Galfalvy et al. BMC Bioinformatics. 2003.
Abstract
Background: Genomic studies of complex tissues pose unique analytical challenges for assessment of data quality, performance of statistical methods used for data extraction, and detection of differentially expressed genes. Ideally, to assess the accuracy of gene expression analysis methods, one needs a set of genes which are known to be differentially expressed in the samples and which can be used as a "gold standard". We introduce the idea of using sex-chromosome genes as an alternative to spiked-in control genes or simulations for assessment of microarray data and analysis methods.
Results: Expression of sex-chromosome genes were used as true internal biological controls to compare alternate probe-level data extraction algorithms (Microarray Suite 5.0 [MAS5.0], Model Based Expression Index [MBEI] and Robust Multi-array Average [RMA]), to assess microarray data quality and to establish some statistical guidelines for analyzing large-scale gene expression. These approaches were implemented on a large new dataset of human brain samples. RMA-generated gene expression values were markedly less variable and more reliable than MAS5.0 and MBEI-derived values. A statistical technique controlling the false discovery rate was applied to adjust for multiple testing, as an alternative to the Bonferroni method, and showed no evidence of false negative results. Fourteen probesets, representing nine Y- and two X-chromosome linked genes, displayed significant sex differences in brain prefrontal cortex gene expression.
Conclusion: In this study, we have demonstrated the use of sex genes as true biological internal controls for genomic analysis of complex tissues, and suggested analytical guidelines for testing alternate oligonucleotide microarray data extraction protocols and for adjusting multiple statistical analysis of differentially expressed genes. Our results also provided evidence for sex differences in gene expression in the brain prefrontal cortex, supporting the notion of a putative direct role of sex-chromosome genes in differentiation and maintenance of sexual dimorphism of the central nervous system. Importantly, these analytical approaches are applicable to all microarray studies that include male and female human or animal subjects.
Figures
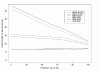
Figure 1
MAS5.0, MBEI and RMA signal variability. The variability in signal intensity measurement obtained with three different probe-level data extraction methods is represented by the lowess curves of the coefficient of variation. The X-axis represent increasing signal intensities, as measured by the percentage of arrays on which this gene is detected as present (% present calls). Presence calls were obtained with MBEI (BA9, n = 39; BA47, n = 36). Note that curves for the two brain areas are very close to each other for all three methods.
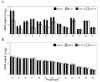
Figure 2
Y-chromosome-linked probesets: male-female expression comparisons. RMA-based averaged values (± STDEV) are displayed. A) Probesets with significant differences in expression levels for male and female samples in BA9 and/or BA47. All male-female comparisons were statistically significant with the exception of #11 in BA9 and # 12 and 13 in BA47 (See also Table 2). Probesets are organized according to order of y-linked genes in Table 2. B) Selected Y-linked probesets without sex-differences. All these genes were detected as ''absent'' by MAS5.0 or MBEI. Signal level represent background estimates. Probesets are: 1, 201909_at; 2, 204409_s_at; 3, 204410_at; 4, 205000_at; 5, 205001_s_at; 6, 206624_at; 7, 206700_s_at; 8, 207063_at; 9, 207246_at; 10, 214983_at; 11, 211149_at; 12, 208067_x_at, 13, 211227_s_at; 14, 214983_at; 15, 217261_at; 16, 217162_at; 17, 221179_at; 18, 211461_at; 19, 209596_at; 20, 210322_x_at; 21, 216376_x_at; 22, 216922_x_at; 23, 211462_s_at; 24, 207909_x_at; 25, 207918_s_at; 26, 207912_s_at.
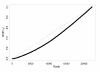
Figure 3
Distribution of the t-tests p-values for sex differences or random group labels. Distribution of the p-values from the t-tests comparing males and females. These p-values are slightly lower than expected from a uniform distribution, representing a mixture distribution of p-values from differentially expressed and not affected genes.
Similar articles
- Comparison of seven methods for producing Affymetrix expression scores based on False Discovery Rates in disease profiling data.
Shedden K, Chen W, Kuick R, Ghosh D, Macdonald J, Cho KR, Giordano TJ, Gruber SB, Fearon ER, Taylor JM, Hanash S. Shedden K, et al. BMC Bioinformatics. 2005 Feb 10;6:26. doi: 10.1186/1471-2105-6-26. BMC Bioinformatics. 2005. PMID: 15705192 Free PMC article. - Correlation test to assess low-level processing of high-density oligonucleotide microarray data.
Ploner A, Miller LD, Hall P, Bergh J, Pawitan Y. Ploner A, et al. BMC Bioinformatics. 2005 Mar 31;6:80. doi: 10.1186/1471-2105-6-80. BMC Bioinformatics. 2005. PMID: 15799785 Free PMC article. - Leveraging two-way probe-level block design for identifying differential gene expression with high-density oligonucleotide arrays.
Barrera L, Benner C, Tao YC, Winzeler E, Zhou Y. Barrera L, et al. BMC Bioinformatics. 2004 Apr 20;5:42. doi: 10.1186/1471-2105-5-42. BMC Bioinformatics. 2004. PMID: 15099405 Free PMC article. - Sex differences in brain expression of X- and Y-linked genes.
Xu J, Disteche CM. Xu J, et al. Brain Res. 2006 Dec 18;1126(1):50-5. doi: 10.1016/j.brainres.2006.08.049. Epub 2006 Sep 7. Brain Res. 2006. PMID: 16962077 Review. - The analysis of microarray data.
Hariharan R. Hariharan R. Pharmacogenomics. 2003 Jul;4(4):477-97. doi: 10.1517/phgs.4.4.477.22744. Pharmacogenomics. 2003. PMID: 12831325 Review.
Cited by
- Comparison of algorithms for the analysis of Affymetrix microarray data as evaluated by co-expression of genes in known operons.
Harr B, Schlötterer C. Harr B, et al. Nucleic Acids Res. 2006 Jan 23;34(2):e8. doi: 10.1093/nar/gnj010. Nucleic Acids Res. 2006. PMID: 16432259 Free PMC article. - Genetic and epigenetic underpinnings of sex differences in the brain and in neurological and psychiatric disease susceptibility.
Qureshi IA, Mehler MF. Qureshi IA, et al. Prog Brain Res. 2010;186:77-95. doi: 10.1016/B978-0-444-53630-3.00006-3. Prog Brain Res. 2010. PMID: 21094887 Free PMC article. Review. - The first decade and beyond of transcriptional profiling in schizophrenia.
Sequeira PA, Martin MV, Vawter MP. Sequeira PA, et al. Neurobiol Dis. 2012 Jan;45(1):23-36. doi: 10.1016/j.nbd.2011.03.001. Epub 2011 Mar 8. Neurobiol Dis. 2012. PMID: 21396449 Free PMC article. Review. - Three clinical variants of gastroesophageal reflux disease form two distinct gene expression signatures.
Ostrowski J, Rubel T, Wyrwicz LS, Mikula M, Bielasik A, Butruk E, Regula J. Ostrowski J, et al. J Mol Med (Berl). 2006 Oct;84(10):872-82. doi: 10.1007/s00109-006-0083-z. Epub 2006 Aug 4. J Mol Med (Berl). 2006. PMID: 16924468 - A cross-laboratory comparison of expression profiling data from normal human postmortem brain.
Mistry M, Pavlidis P. Mistry M, et al. Neuroscience. 2010 May 5;167(2):384-95. doi: 10.1016/j.neuroscience.2010.01.016. Epub 2010 Feb 4. Neuroscience. 2010. PMID: 20138973 Free PMC article.
References
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH064168/MH/NIMH NIH HHS/United States
- T32 MH020004/MH/NIMH NIH HHS/United States
- MH62185/MH/NIMH NIH HHS/United States
- P50 MH062185/MH/NIMH NIH HHS/United States
- F32MH63559/MH/NIMH NIH HHS/United States
- K01 MH067721/MH/NIMH NIH HHS/United States
- R01 MH40210/MH/NIMH NIH HHS/United States
- F32 MH063559/MH/NIMH NIH HHS/United States
- R01 MH040210/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources