Rationally designed insulator-like elements can block enhancer action in vitro - PubMed (original) (raw)
Rationally designed insulator-like elements can block enhancer action in vitro
Vladimir A Bondarenko et al. EMBO J. 2003.
Abstract
Insulators are DNA sequences that are likely to be involved in formation of chromatin domains, functional units of gene expression in eukaryotes. Insulators can form domain boundaries and block inappropriate action of regulatory elements (such as transcriptional enhancers) in eukaryotic nuclei. Using an in vitro system supporting enhancer action over a large distance, the enhancer-blocking insulator activity has been recapitulated in a highly purified system. The insulator-like element was constructed using a sequence-specific DNA-binding protein making stable DNA loops (lac repressor). The insulation was entirely dependent on formation of a DNA loop that topologically isolates the enhancer from the promoter. This rationally designed, inducible insulator-like element recapitulates many key properties of eukaryotic insulators observed in vivo. The data suggest novel mechanisms of enhancer and insulator action.
Figures
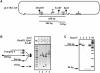
Fig. 1. The enhancer cannot _cis_-activate the glnAp2 promoter localized within a topologically isolated DNA loop. (A) The branch collision mechanism. This mechanism was proposed to explain the effect of DNA supercoiling on the rate of communication between DNA regions on supercoiled DNA. Frequent collisions between branches formed on supercoiled DNA could facilitate communication between the enhancer and promoter localized on different branches of the same DNA molecule. The enhancer and promoter are indicated by black and white circles, respectively. (B) LacI-induced DNA loop formation topologically isolates the enhancer from the promoter. When spaced by 2.5 kb, the enhancer (E) and promoter (P) probably communicate by slithering on supercoiled DNA (structures 1 and 2). The slithering model suggests that intertwined DNA helixes can slide relative to each other on supercoiled DNA at a high rate; sliding greatly increases the probability of enhancer–promoter collision. LacI tetramer (R) can simultaneously bind two lac operators positioned upstream and downstream of the promoter (O1 and O2), form a DNA loop (structure 3), and thus could topologically isolate the enhancer from the promoter. (C) LacI-induced DNA loop formation greatly inhibits enhancer-dependent transcription. The experimental strategy is outlined at the top. Plasmid containing two lac operators localized 0.36 kb upstream and 0.27 kb downstream of the promoter [pLY10(1-1)S, upper panel] and control plasmid not containing lac operators (pLY10, lower panel) were transcribed in the presence or in the absence of wt LacI or the R3 mutant. R3 mutant binds DNA well but cannot form DNA loops. In some cases, LacI was added after formation of the open complexes (lanes 4 and 10). The distances between key regulatory elements are shown in italics. RPo, open initiation complex. Other designations are as in (B). M, labeled pBR322-MspI markers. Note that, although LacI can present a weak road-block to transcript elongation (Oehler et al., 1990), the block was not detected on the templates used in this work. (D) LacI and the R3 mutant LacI are quantitatively bound to their binding sites. Analysis by native PAGE. Aliquots of transcription reactions described in (C), lanes 2, 3, 5 and 6, containing LacI or R3, respectively, but not containing plasmid DNA were incubated in the presence of labeled double-stranded oligonucleotide containing an ‘ideal’ lac operator. Mobilities of corresponding complexes in the gel are indicated.
Fig. 2. Topological isolation of the enhancer from the promoter reduces the rate of enhancer–promoter communication. (A) Transcription assay for analysis of the effect of LacI on enhancer–promoter communication. The pLY10(1-1)S supercoiled template was pre-incubated with all components of the transcription machinery, and then incubated with or without LacI. Then ATP was added to allow enhancer–promoter communication. The rate of open complex formation was measured in a single-round transcription assay. NtrC activator octamer bound to the enhancer (E) and RNA polymerase bound to the promoter (P) are shown. Other designations are as in Figure 1C. (B) Analysis of labeled specific transcripts by denaturing PAGE. (C) The rate of enhancer– promoter communication is strongly decreased in the presence of LacI. DNA bands containing specific transcripts (B) were quantified and plotted as a function of time.
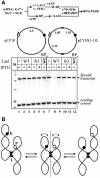
Fig. 3. LacI interacts with two lac operators and forms a stable loop on supercoiled DNA. (A) Partial restriction map of the pLY10(1-1)S plasmid. If LacI interacts with both lac operators (O1 and O2) on supercoiled DNA, two DNA fragments (84 and 566 bp) generated after digestion with restriction enzymes _Hin_dIII, _Eco_RI and _Kpn_I are expected to be bound to LacI and migrate as a single DNA–protein complex in a native gel. (B) LacI-induced DNA loop formation on supercoiled DNA: analysis of DNA–protein complexes. Supercoiled pLY10(1-1)S template was pre-incubated with or without LacI, then digested by different combinations of restriction enzymes and analyzed in a native agarose gel. The expected products of digestion are shown on the left. Different bands in the gel are arbitrarily numbered. M, 100 bp DNA ladder (NEB). (C) LacI-induced DNA loop formation on supercoiled DNA: analysis of DNA composition of different DNA–protein complexes. DNA was purified from different bands [1–4 in (B)] and analyzed by PAGE. M, 100 bp DNA ladder (NEB).
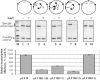
Fig. 4. Topological isolation of the enhancer from the promoter is required for insulation. Various constructs (upper panel), all containing the enhancer and promoter spaced by 2.5 kb but different numbers of ‘ideal’ lac operators positioned differently were transcribed in the presence or in the absence of LacI. The arrows between lac operators indicate the expected LacI-induced DNA loops. Two alternative loops formed on the pLY10(2-1) plasmid are indicated (arrows 1 and 2); other designations are as in Figure 1C. All constructs were transcribed in the same experiment; transcripts were analyzed by denaturing PAGE (middle panel). Equal amounts of a labeled DNA fragment were added to all samples immediately after termination of transcription (loading control). The histogram (lower panel) shows the ratio of the amounts of specific transcripts accumulated in the presence and in the absence of LacI. The data are averages of at least three experiments (standard deviations are indicated).
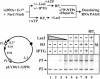
Fig. 5. LacI-induced DNA loop topologically isolates the enhancer from the promoter but does not inactivate them. The pLY10(1-1)2PR plasmid (upper panel) containing two identical glnAp2 promoters (P1 and P2) under the control of one enhancer and two lac operators flanking promoter P1 was transcribed in the presence or in the absence of LacI and IPTG (see the diagram at the top). The regions transcribed from the P1 and P2 promoters are 401 and 309 nt long, respectively. The transcripts were analyzed by denaturing PAGE (lower panel). The template concentration was 3 nM and lac repressor was added to the following concentrations: lane 4, 2 nM; lane 3, 4 nM; lane 2, 6 nM and lanes 1 and 6–9, 8 nM. Other designations are as in Figure 1C.
Fig. 6. Two lac operators can inhibit enhancer-dependent transcription over a large distance. (A) Two lac operators positioned 3.5 kb from each other can inhibit enhancer-dependent transcription. Plasmid containing two lac operators localized 3.7 kb from each other and at least 1.4 kb from the promoter [pLY10(1-1)L] and control plasmid not containing lac operators (pLY10) were transcribed in the presence or in the absence of LacI or the R3 mutant (see diagram at the top). A single-round assay was used. In some cases, LacI was added after formation of the open complexes (lanes 6 and 12). Labeled transcripts were analyzed in a denaturing gel. Labeled DNA fragment was added to all samples immediately after termination of transcription (loading control). Other designations are as in Figure 1C. (B) Slithering between two topologically isolated domains is impossible. Designations are as in Figure 1B. Binding of LacI tetramer to two lac operators results in separation of plasmid DNA into two topological domains. Slithering is only possible within each domain but not between them. Placing promoter and enhancer in topologically isolated domains prevents communication between them by slithering on supercoiled DNA.
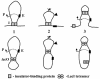
Fig. 7. Similarities between action of natural insulators in vivo and lac operators in vitro. An enhancer (E) and promoter (P) can efficiently communicate when flanked by insulators (I) or lac operators (_lac_O, structure 1). Insertion of an insulator or lacO between an enhancer and promoter may result in formation of a DNA loop and topological isolation of the enhancer from the promoter that prevents enhancer– promoter communication (structure 2). Incorporation of a second insulator between the enhancer and promoter (indicated by arrowhead in structure 2) may result in formation of a competitive DNA loop (structure 3). As a result, the enhancer and promoter would reside in the same topological domain and can efficiently communicate with each other. Many enhancer-blocking properties of insulators and lac operators can be rationalized in the framework of the slithering model of enhancer action (see Discussion). Note that only interactions between two eukaryotic insulators are shown (upper panel) while in vivo insulators form large ‘insulator bodies’ (Gerasimova and Corces, 1998; Gerasimova et al., 2000; Ishii et al., 2002).
Similar articles
- Communication over a large distance: enhancers and insulators.
Bondarenko VA, Liu YV, Jiang YI, Studitsky VM. Bondarenko VA, et al. Biochem Cell Biol. 2003 Jun;81(3):241-51. doi: 10.1139/o03-051. Biochem Cell Biol. 2003. PMID: 12897858 Review. - [Mechanisms of distant enhancer action on DNA and in chromatin].
Studitskiĭ VM. Studitskiĭ VM. Mol Biol (Mosk). 2009 Mar-Apr;43(2):204-14. Mol Biol (Mosk). 2009. PMID: 19425490 Review. Russian. - Functional characterization of the enhancer blocking element of the sea urchin early histone gene cluster reveals insulator properties and three essential cis-acting sequences.
Melfi R, Palla F, Di Simone P, Alessandro C, Calì L, Anello L, Spinelli G. Melfi R, et al. J Mol Biol. 2000 Dec 15;304(5):753-63. doi: 10.1006/jmbi.2000.4273. J Mol Biol. 2000. PMID: 11124024 - Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo.
Cai H, Levine M. Cai H, et al. Nature. 1995 Aug 10;376(6540):533-6. doi: 10.1038/376533a0. Nature. 1995. PMID: 7637789 - Insulators: exploiting transcriptional and epigenetic mechanisms.
Gaszner M, Felsenfeld G. Gaszner M, et al. Nat Rev Genet. 2006 Sep;7(9):703-13. doi: 10.1038/nrg1925. Epub 2006 Aug 15. Nat Rev Genet. 2006. PMID: 16909129 Review.
Cited by
- Mechanism of chromosomal boundary action: roadblock, sink, or loop?
Gohl D, Aoki T, Blanton J, Shanower G, Kappes G, Schedl P. Gohl D, et al. Genetics. 2011 Mar;187(3):731-48. doi: 10.1534/genetics.110.123752. Epub 2010 Dec 31. Genetics. 2011. PMID: 21196526 Free PMC article. - Probability of the site juxtaposition determines the rate of protein-mediated DNA looping.
Polikanov YS, Bondarenko VA, Tchernaenko V, Jiang YI, Lutter LC, Vologodskii A, Studitsky VM. Polikanov YS, et al. Biophys J. 2007 Oct 15;93(8):2726-31. doi: 10.1529/biophysj.107.111245. Epub 2007 Jun 15. Biophys J. 2007. PMID: 17573434 Free PMC article. - Selective interactions between diverse STEs organize the ANT-C Hox cluster.
Li M, Ma Z, Roy S, Patel SK, Lane DC, Duffy CR, Cai HN. Li M, et al. Sci Rep. 2018 Oct 11;8(1):15158. doi: 10.1038/s41598-018-33588-4. Sci Rep. 2018. PMID: 30310129 Free PMC article. - Enhancer-promoter communication is regulated by insulator pairing in a Drosophila model bigenic locus.
Maksimenko O, Golovnin A, Georgiev P. Maksimenko O, et al. Mol Cell Biol. 2008 Sep;28(17):5469-77. doi: 10.1128/MCB.00461-08. Epub 2008 Jun 23. Mol Cell Biol. 2008. PMID: 18573869 Free PMC article. - Chromatin boundary elements organize genomic architecture and developmental gene regulation in Drosophila Hox clusters.
Ma Z, Li M, Roy S, Liu KJ, Romine ML, Lane DC, Patel SK, Cai HN. Ma Z, et al. World J Biol Chem. 2016 Aug 26;7(3):223-30. doi: 10.4331/wjbc.v7.i3.223. World J Biol Chem. 2016. PMID: 27621770 Free PMC article. Review.
References
- Bagga R. and Emerson,B.M. (1997) An HMG I/Y-containing repressor complex and supercoiled DNA topology are critical for long-range enhancer-dependent transcription in vitro. Genes Dev., 11, 629–639. - PubMed
- Barkley M.D., Riggs,A.D., Jobe,A. and Burgeois,S. (1975) Interaction of effecting ligands with lac repressor and repressor-operator complex. Biochemistry, 14, 1700–1712. - PubMed
- Barry J.K. and Matthews,K.S. (1997) Ligand-induced conformational changes in lactose repressor: a fluorescence study of single tryptophan mutants. Biochemistry, 36, 15632–15642. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources