Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins - PubMed (original) (raw)
Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins
Nathalie de Parseval et al. J Virol. 2003 Oct.
Abstract
Sequences of retroviral origin occupy approximately 8% of the human genome. Most of these "retroviral" genes have lost their coding capacities since their entry into our ancestral genome millions of years ago, but some reading frames have remained open, suggesting positive selection. The complete sequencing of the human genome allowed a systematic search for retroviral envelope genes containing an open reading frame and resulted in the identification of 16 genes that we have characterized. We further showed, by quantitative reverse transcriptase PCR using specifically devised primers which discriminate between coding and noncoding elements, that all 16 genes are expressed in at least some healthy human tissues, albeit at highly different levels. All envelope genes disclose significant expression in the testis, three of them have a very high level of expression in the placenta, and a fourth is expressed in the thyroid. Besides their primary role as key molecules for viral entry, the envelope genes of retroviruses can induce cell-cell fusion, elicit immunosuppressive effects, and even protect against infection, and as such, endogenous retroviral envelope proteins have been tentatively identified in several reports as being involved in both normal and pathological processes. The present study provides a comprehensive survey of candidate genes and tools for a precise evaluation of their involvement in these processes.
Figures
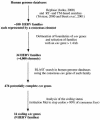
FIG. 1.
Flow chart for the identification of the 16 coding envelope genes of the human genome (see Results).
FIG. 2.
Structural organization and characteristic features of the 16 identified HERV envelope proteins. Hydrophobicity profiles and characteristic domains of the 16 HERV envelope proteins (the six HERV-K HML2 envelope proteins that are almost identical are represented by a single protein) and of the Moloney murine leukemia virus envelope protein (EnvMo) are shown. The black frames delineate the ORFs, and the gray frames at the end of the R and F(c)2 envelope proteins represent the short reading frames present downstream of the stop codon. Hatched areas correspond to hydrophobic regions associated with the fusion peptide and the transmembrane region, respectively; the CWLC motifs (consensus, C-X-X-C) and the proteolytic cleavage sites (consensus, R/K-X-R/K-R) are represented by light and dark gray vertical bars, respectively, and their sequences are indicated on the right; the putative immunosuppressive domain is represented with a gray box.
FIG. 3.
SDS-PAGE analysis of the radiolabeled proteins produced upon direct in vitro transcription-translation assays of the amplification products of the coding retroviral envelope genes obtained from the corresponding BAC clones. Arrowheads point to the translation products of the expected size for each envelope gene. Bands of higher molecular weight are observed for EnvF(c)2, most probably associated with the presence of a frameshift signal at the env gene 3′ end (3).
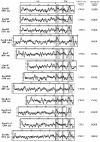
FIG. 4.
Transcript levels of the coding retroviral envelope genes of the human genome in a panel of 19 healthy human tissues, as determined by real-time quantitative PCR. Each value is expressed as the mean ± standard deviation relative to the value for a reference control plasmid assayed in each real-time PCR experiment (see Materials and Methods), after correction for total RNA content in each tissue extract (using the 18S transcript as an internal control). The code for the logarithmic gray scale is indicated on the right. PBL, peripheral blood lymphocytes.
Similar articles
- Comprehensive search for intra- and inter-specific sequence polymorphisms among coding envelope genes of retroviral origin found in the human genome: genes and pseudogenes.
de Parseval N, Diop G, Blaise S, Helle F, Vasilescu A, Matsuda F, Heidmann T. de Parseval N, et al. BMC Genomics. 2005 Sep 9;6:117. doi: 10.1186/1471-2164-6-117. BMC Genomics. 2005. PMID: 16150157 Free PMC article. - Capture of a Hyena-Specific Retroviral Envelope Gene with Placental Expression Associated in Evolution with the Unique Emergence among Carnivorans of Hemochorial Placentation in Hyaenidae.
Funk M, Cornelis G, Vernochet C, Heidmann O, Dupressoir A, Conley A, Glickman S, Heidmann T. Funk M, et al. J Virol. 2019 Feb 5;93(4):e01811-18. doi: 10.1128/JVI.01811-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463979 Free PMC article. - Human endogenous retroviruses: from infectious elements to human genes.
de Parseval N, Heidmann T. de Parseval N, et al. Cytogenet Genome Res. 2005;110(1-4):318-32. doi: 10.1159/000084964. Cytogenet Genome Res. 2005. PMID: 16093684 Review. - [Envelope capture by retroviruses].
Kim FJ, Manel N, Battini JL, Sitbon M. Kim FJ, et al. Med Sci (Paris). 2004 Oct;20(10):876-81. doi: 10.1051/medsci/20042010876. Med Sci (Paris). 2004. PMID: 15461964 Review. French. - From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation.
Dupressoir A, Lavialle C, Heidmann T. Dupressoir A, et al. Placenta. 2012 Sep;33(9):663-71. doi: 10.1016/j.placenta.2012.05.005. Epub 2012 Jun 12. Placenta. 2012. PMID: 22695103 Review.
Cited by
- The Relationship between HERV, Interleukin, and Transcription Factor Expression in ZIKV Infected versus Uninfected Trophoblastic Cells.
Costa ALD, Prieto-Oliveira P, Duarte-Barbosa M, Andreata-Santos R, Peter CM, Prolo de Brito T, Antoneli F, Durães-Carvalho R, Briones MRS, Maricato JT, Zanotto PMA, Jacob Machado D, Janini LMR. Costa ALD, et al. Cells. 2024 Sep 5;13(17):1491. doi: 10.3390/cells13171491. Cells. 2024. PMID: 39273061 Free PMC article. - Recombinant origin and interspecies transmission of a HERV-K(HML-2)-related primate retrovirus with a novel RNA transport element.
Williams ZH, Imedio AD, Gaucherand L, Lee DC, Mostafa SM, Phelan JP, Coffin JM, Johnson WE. Williams ZH, et al. Elife. 2024 Jul 22;13:e80216. doi: 10.7554/eLife.80216. Elife. 2024. PMID: 39037763 Free PMC article. - Galectin-1 Modulates the Fusogenic Activity of Placental Endogenous Retroviral Envelopes.
Toudic C, Maurer M, St-Pierre G, Xiao Y, Bannert N, Lafond J, Rassart É, Sato S, Barbeau B. Toudic C, et al. Viruses. 2023 Dec 16;15(12):2441. doi: 10.3390/v15122441. Viruses. 2023. PMID: 38140682 Free PMC article. - Virus-Induced Cell Fusion and Syncytia Formation.
Xie M. Xie M. Results Probl Cell Differ. 2024;71:283-318. doi: 10.1007/978-3-031-37936-9_14. Results Probl Cell Differ. 2024. PMID: 37996683 - Syncytin-1, syncytin-2 and suppressyn in human health and disease.
Priščáková P, Svoboda M, Feketová Z, Hutník J, Repiská V, Gbelcová H, Gergely L. Priščáková P, et al. J Mol Med (Berl). 2023 Dec;101(12):1527-1542. doi: 10.1007/s00109-023-02385-6. Epub 2023 Oct 19. J Mol Med (Berl). 2023. PMID: 37855856 Free PMC article. Review.
References
- Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. - PubMed
- Benit, L., A. Calteau, and T. Heidmann. 2003. Characterization of the low copy HERV-Fc family: evidence for recent integrations in primates of elements with coding envelope genes. Virology 312:159-168. - PubMed
- Best, S., P. R. Le Tissier, and J. P. Stoye. 1997. Endogenous retroviruses and the evolution of resistance to retroviral infection. Trends Microbiol. 5:313-318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases