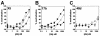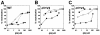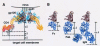Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1 - PubMed (original) (raw)
. 2003 Oct;77(19):10557-65.
doi: 10.1128/jvi.77.19.10557-10565.2003.
Pascal Poignard, Aarti Raja, Michael B Zwick, Karla Delgado, Michael Franti, James Binley, Veronique Vivona, Christoph Grundner, Chih-Chin Huang, Miro Venturi, Christos J Petropoulos, Terri Wrin, Dimiter S Dimitrov, James Robinson, Peter D Kwong, Richard T Wyatt, Joseph Sodroski, Dennis R Burton
Affiliations
- PMID: 12970440
- PMCID: PMC228502
- DOI: 10.1128/jvi.77.19.10557-10565.2003
Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1
Aran F Labrijn et al. J Virol. 2003 Oct.
Abstract
Anti-human immunodeficiency virus type 1 (HIV-1) antibodies whose binding to gp120 is enhanced by CD4 binding (CD4i antibodies) are generally considered nonneutralizing for primary HIV-1 isolates. However, a novel CD4i-specific Fab fragment, X5, has recently been found to neutralize a wide range of primary isolates. To investigate the precise nature of the extraordinary neutralizing ability of Fab X5, we evaluated the abilities of different forms (immunoglobulin G [IgG], Fab, and single-chain Fv) of X5 and other CD4i monoclonal antibodies to neutralize a range of primary HIV-1 isolates. Our results show that, for a number of isolates, the size of the neutralizing agent is inversely correlated with its ability to neutralize. Thus, the poor ability of CD4i-specific antibodies to neutralize primary isolates is due, at least in part, to steric factors that limit antibody access to the gp120 epitopes. Studies of temperature-regulated neutralization or fusion-arrested intermediates suggest that the steric effects are important in limiting the binding of IgG to the viral envelope glycoproteins after HIV-1 has engaged CD4 on the target cell membrane. The results identify hurdles in using CD4i epitopes as targets for antibody-mediated neutralization in vaccine design but also indicate that the CD4i regions could be efficiently targeted by small molecule entry inhibitors.
Figures
FIG. 1.
Neutralization of HIV-1JR-CSF by CD4i antibodies and antibody fragments. Neutralization titers of whole antibodies (•), Fab fragments (○), and scFv fragments (▪) to HIV-1JR-CSF were determined in a pseudotyped luciferase-based neutralization assay with U87.CD4.CCR5 cells. Datum points are the means of triplicates ± the standard error of the mean (SEM).
FIG. 2.
Binding curves of CD4i antibodies and antibody fragments to monomeric gp120JR-CSF as determined by ELISA. Titers of whole antibodies (A) and Fab fragments (B) to gp120JR-CSF were determined in the presence (solid symbols) or absence (open symbols) of saturating amounts of sCD4. Symbols: circles, 17b; squares, 48d; triangles, X5. Datum points are the means of at least two separate experiments ± the standard deviation.
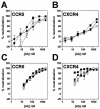
FIG. 3.
Neutralization of dualtropic HIV-1 by MAb X5 whole IgG and antibody fragments. Titers of whole antibody (•), Fab fragment (○) and scFv fragment (▪) to HIV-189.6 (A and B) and HIV-1JR-CSF/IR (C and D) were determined in a pseudotyped luciferase-based neutralization assay with U87.CD4 cells expressing either CCR5 (A and C) or CXCR4 (B and D). Datum points are the means of triplicates ± the SEM.
FIG. 4.
Pre- and postattachment neutralization of HIV-1ADA and HIV-1ADAΔV1V2 by MAb 17b whole antibody and antibody fragments. Neutralization titers of whole antibody (•), Fab fragment (○), and scFv fragment (▪) to HIV-1ADA (A) and HIV-1ADAΔV1V2 (B and C) were determined in a pseudotyped luciferase-based neutralization assay with Cf2Th cells expressing CD4 and CCR5. In these settings pre- and postattachment neutralizations were measured simultaneously (A and B) by preincubating virus and antibody fragments at 37°C prior to addition to cells. (C) Postattachment neutralization was measured exclusively by preincubating virus and cells at 4°C, washing the cells, and adding antibody fragments before the temperature was increased to 37°C.
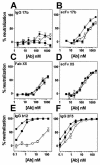
FIG. 5.
Standard and postattachment neutralization of SOSJR-FL and WTJR-FL by CD4i whole antibodies and antibody fragments. Antibodies 17b (IgG [A] and scFv [B]), X5 (Fab [C] and scFv [D]), b12 (IgG [E]), and 2F5 (IgG [F]) were tested in pseudotyped luciferase-based neutralization assays, under standard assay conditions against WTJR-FL (▪) and SOSJR-FL, and in a postattachment format against SOSJR-FL (○). Datum points are the means of triplicates ± the SEM.
FIG. 6.
Steric restrictions on CD4i antibody neutralization after CD4 attachment. (A) HIV-1 viral spike attached to cell surface CD4. The gp120 trimer (brown) (27) is shown with the threefold axis oriented perpendicular to the target cell surface. Cα worm representations of gp120 and four-domain CD4 are shown in brown and yellow, respectively. N-linked carbohydrate on gp120 is shown in blue. The flexibility between the second and third extracellular CD4 domains allows the observed angle between the membrane-distal (D1D2) and membrane-proximal (D3D4) domains to vary by up to 10° (distinct crystal structure conformations are shown in red and yellow) (58). The distance between the membrane-proximal portion of gp120 and the bottom of the crystallographically ordered CD4 is ∼45 Å. Nine amino acids (shown in gray) occur between the last ordered crystallographic residue (position 363) and the transmembrane domain (starting at position 373), potentially increasing the gp120-target cell membrane distance by 20 Å. There is considerable uncertainty in the degree of flexibility between the second and third extracellular domains of CD4. If the interdomain rotation is increased by 35° (gray), the gp120-target cell membrane distance would increase by an additional 20 Å, resulting in a total gp120-transmembrane distance of up to 85 Å. (B) Dimensions of Fv, Fab, and IgG portions of 17b interacting with gp120. Both gp120 (brown) and antibody fragments (purple) are shown in the Cα worm representation. The gp120 molecule is depicted in precisely the same orientation as the front left protomer in panel A, with the 17b portion oriented by superimposing the X-ray structure (pdb accession number 1G9 M [25]) of the gp120 ternary complex with D1D2 and 17b Fab complex. The length of the 17b Fv along the trimer threefold axis is ∼40 Å, and the length of the Fab is ∼60 Å, with the dimensions measured for the distance between the target membrane-proximal portions of gp120 and 17b. For IgGs, considerable flexibility is found in the hinge region connecting Fab and Fc portions of the IgG. Superimposing the crystal structure of a full-length human antibody (45) onto 17b gives dimensions of 115 Å (shown here) and 140 Å (not shown) for the two alternative Fab superpositions. The figures were generated with GRASP (34).
Similar articles
- Unique binding modes for the broad neutralizing activity of single-chain variable fragments (scFv) targeting CD4-induced epitopes.
Tanaka K, Kuwata T, Alam M, Kaplan G, Takahama S, Valdez KPR, Roitburd-Berman A, Gershoni JM, Matsushita S. Tanaka K, et al. Retrovirology. 2017 Sep 22;14(1):44. doi: 10.1186/s12977-017-0369-y. Retrovirology. 2017. PMID: 28938888 Free PMC article. - Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1.
Roben P, Moore JP, Thali M, Sodroski J, Barbas CF 3rd, Burton DR. Roben P, et al. J Virol. 1994 Aug;68(8):4821-8. doi: 10.1128/JVI.68.8.4821-4828.1994. J Virol. 1994. PMID: 7518527 Free PMC article. - Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes.
Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X, Binley JM, Zhang MY, Sidorov IA, Broder CC, Robinson J, Parren PW, Burton DR, Dimitrov DS. Moulard M, et al. Proc Natl Acad Sci U S A. 2002 May 14;99(10):6913-8. doi: 10.1073/pnas.102562599. Epub 2002 May 7. Proc Natl Acad Sci U S A. 2002. PMID: 11997472 Free PMC article. - Novel approaches for identification of broadly cross-reactive HIV-1 neutralizing human monoclonal antibodies and improvement of their potency.
Zhang MY, Dimitrov DS. Zhang MY, et al. Curr Pharm Des. 2007;13(2):203-12. doi: 10.2174/138161207779313669. Curr Pharm Des. 2007. PMID: 17269928 Review. - HIV coreceptors: from discovery and designation to new paradigms and promise.
Alkhatib G, Berger EA. Alkhatib G, et al. Eur J Med Res. 2007 Oct 15;12(9):375-84. Eur J Med Res. 2007. PMID: 17933717 Review.
Cited by
- Chimeric HIV-1 envelope glycoproteins with potent intrinsic granulocyte-macrophage colony-stimulating factor (GM-CSF) activity.
Isik G, van Montfort T, Boot M, Cobos Jiménez V, Kootstra NA, Sanders RW. Isik G, et al. PLoS One. 2013;8(4):e60126. doi: 10.1371/journal.pone.0060126. Epub 2013 Apr 2. PLoS One. 2013. PMID: 23565193 Free PMC article. - Elicitation of neutralizing antibodies directed against CD4-induced epitope(s) using a CD4 mimetic cross-linked to a HIV-1 envelope glycoprotein.
Dey AK, Burke B, Sun Y, Sirokman K, Nandi A, Hartog K, Lian Y, Geonnotti AR, Montefiori D, Franti M, Martin G, Carfi A, Kessler P, Martin L, Srivastava IK, Barnett SW. Dey AK, et al. PLoS One. 2012;7(1):e30233. doi: 10.1371/journal.pone.0030233. Epub 2012 Jan 24. PLoS One. 2012. PMID: 22291921 Free PMC article. - Local conformational stability of HIV-1 gp120 in unliganded and CD4-bound states as defined by amide hydrogen/deuterium exchange.
Kong L, Huang CC, Coales SJ, Molnar KS, Skinner J, Hamuro Y, Kwong PD. Kong L, et al. J Virol. 2010 Oct;84(19):10311-21. doi: 10.1128/JVI.00688-10. Epub 2010 Jul 21. J Virol. 2010. PMID: 20660185 Free PMC article. - Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles.
Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S. Gorny MK, et al. J Virol. 2005 Apr;79(8):5232-7. doi: 10.1128/JVI.79.8.5232-5237.2005. J Virol. 2005. PMID: 15795308 Free PMC article.
References
- Barbas, C. F., III, D. R. Burton, J. K. Scott, and G. J. Silverman. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. - PMC - PubMed
- Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI42848/AI/NIAID NIH HHS/United States
- R37 AI024755/AI/NIAID NIH HHS/United States
- R01 AI041851/AI/NIAID NIH HHS/United States
- AI31783/AI/NIAID NIH HHS/United States
- R01 AI031783/AI/NIAID NIH HHS/United States
- AI39420/AI/NIAID NIH HHS/United States
- R01 AI039420/AI/NIAID NIH HHS/United States
- AI41851/AI/NIAID NIH HHS/United States
- P30 AI042848/AI/NIAID NIH HHS/United States
- R37 AI033292/AI/NIAID NIH HHS/United States
- R01 AI033292/AI/NIAID NIH HHS/United States
- R01 AI045357/AI/NIAID NIH HHS/United States
- AI33292/AI/NIAID NIH HHS/United States
- AI49566/AI/NIAID NIH HHS/United States
- AI45357/AI/NIAID NIH HHS/United States
- AI24755/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials