The role of the 3' untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells - PubMed (original) (raw)
The role of the 3' untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells
J Sylvestre et al. Mol Biol Cell. 2003 Sep.
Abstract
We recently demonstrated, using yeast DNA microarrays, that mRNAs of polysomes that coisolate with mitochondria code for a subset of mitochondrial proteins. The majority of these mRNAs encode proteins of prokaryotic origin. Herein, we show that a similar association occurs between polysomes and mitochondria in human cells. To determine whether mRNA transport machinery is conserved from yeast to human cells, we examined the subcellular localization of human OXA1 mRNA in yeast. Oxa1p is a key component in the biogenesis of mitochondrial inner membrane and is conserved from bacteria to eukaryotic organelles. The expression of human OXA1 cDNA partially restores the respiratory capacity of yeast oxa1- cells. In this study, we demonstrate that 1) OXA1 mRNAs are remarkably enriched in mitochondrion-bound polysomes purified from yeast and human cells; 2) the presence of the human OXA1 3' untranslated region (UTR) is required for the function of the human Oxa1p inside yeast mitochondria; and 3) the accurate sorting of the human OXA1 mRNA to the vicinity of yeast mitochondria is due to the recognition by yeast proteins of the human 3' UTR. Therefore, it seems that the recognition mechanism of OXA1 3' UTR is conserved throughout evolution and is necessary for Oxa1p function.
Figures
Figure 1.
Subellular distribution of mRNAs encoding mitochondrial proteins in HeLa cells. (A) Northern blots were performed with RNA prepared from free polysomes (F-P) and mitochondrion-bound polysomes (M-P) by using probes for different genes encoding mitochondrial proteins (Table 1). At the bottom, methylene blue staining of the ribosomal RNAs is shown. The exposure times of the autoradiograms were ∼14 h at –80°C by using Amersham intensifying screens for all the probes except the 5.6-kb mtDNA, which reveals the ND5, ND4, and COX3 genes, the sizes of these three transcripts are 1.9, 1.45, and 1 kb, respectively, and required an exposure time of only 1 h. (B) Quantifications of the hybridization signals were made using the PhosphorImager system and TINA software. The signal obtained for an individual transcript in the M-P fraction was normalized with the COX3 signal. Normalization for the signal obtained in the F-P fraction was performed using the IRP1 signal. For a given mRNA, addition of both signals after normalization was considered as 100%. The approximate percentage is a mean of four independent biochemical purification experiments, and the Northern blot analyses were performed twice for each polysomal preparation. (C) RT-PCR analysis were performed with 200 ng of RNA purified from M-P and F-P by using specific oligonucleotides within COX10, COX6c, and NDUFV1 ORFs (Table 1), the size of each amplified product is shown at the bottom. The four independent polysomal RNA purifications were subjected twice to RT-PCR analyses.
Figure 2.
OXA1 mRNA localizes to the vicinity of mitochondria in both yeast and human cells. RT-PCR analyses were performed with RNAs purified from mitochondrion-bound polysomes (M-P), free cytoplasmic polysomes (F-P) in yeast and HeLa cells. Specific oligonucleotides were chosen inside each coding region for human OXA1, ferritin, and ATP5b genes, yeast OXA1, ATP2, ACT1, and COX3 genes (Table 1) to perform RT-PCR analyses. Ten percent of the amplified products were subjected to electrophoresis; the size of each amplified product is shown at the bottom.
Figure 3.
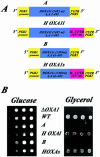
Human OXA1 mRNAs expressed in yeast cells are found associated with polysomes bound to the mitochondrial surface. Yeast oxa1_– mutant cells (Δ_oxa1, strain NBT1) were transformed with plasmids directing the expression of either the full-length Oxa1 protein (HOXA1l) or the N-terminal truncated form (HOXA1s). (A) Schematic representation of the two plasmids. The expression of both human proteins is under the regulation of the yeast PGK1 promoter. Both constructions share at their 3′ extremities 237 nucleotides of the human OXA1 3′ UTR sequence and the yeast PGK1 3′ UTR. (B) The respiratory growth of the transformants and wild-type cells (WT) was examined on glycerol medium at 28°C. The image represents an incubation of 3 d. (C) RT-PCR analysis were performed with RNAs from free cytoplasmic polysomes (F-P), mitochondrion-bound polysomes (M-P) purified from WT cells and _oxa1_– cells expressing either the HOXA1l or the HOXA1s plasmid. Specific oligonucleotides were chosen inside the coding region for human OXA1, yeast OXA1, ATP2, and COX6 sequences (Table 1). Ten percent of each amplified product was subjected to electrophoresis. (D) Quantifications were made using the TINA software. For the M-P polysomes, the OXA1 RT-PCR product signal was normalized using the ATP2 signal. For the F-P polysomes, the OXA1's RT-PCR signal was normalized using the COX6 signal. The experiments were performed with four independent polysomal RNA purifications, and each polysomal preparation was subjected to RT-PCR analysis twice.
Figure 4.
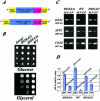
The human OXA1 3′ UTR is essential to rescue the respiratory function of yeast oxa1 mutant cells. (A) Schematic representation of the four constructions. The complete human OXA1 3′UTR was deleted from the original constructions directing the expression of either the full-length human Oxa1 protein (HOXA1l) or the N-terminal truncated version (HOXA1s) to give HOXA1/A et HOXA1s/B. (B) Yeast oxa1_-mutant cells (Δ_oxa1, strain NBT1) were transformed with either one of the four plasmids (A) and the respiratory capacity of the transformed cells was tested by serially diluting cells at OD600 of 2 and plating in both glucose medium or glycerol medium. Images represent an incubation of 4 d at 28°C.
Figure 5.
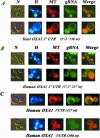
Imaging fluorescent RNAs in living yeast cells. Coexpression of CP-GFP plasmid and reporter RNAs, leads a GFP-labeled RNA, which was visualized using fluorescence microscopy techniques. Cells were grown in 2% galactose medium or incubated in a 2% glycerol medium to detect cells in which mitochondria consist of a branched tubular network and visualized at early log phase. gRNA indicates the green RNA labeling, H the Hoechst staining, N the cells photographed with Nomarski optics, and MT cells labeled with the MitoTracker dye. The merge at the right of the figure represents the superposition of both green RNA fluorescence and mitochondrial labeling with MitoTracker. (A) Reporter RNA with the yeast OXA1 3′ UTR of 158 nucleotides in length cloned in the coding orientation. Top line, CP-GFP expression was induced for 2 h in 2% galactose medium devoid of methionine Bottom line, CP-GFP expression was induced for2hin2% glycerol medium devoid of methionine. (B) Reporter RNA with the human 3′ UTR of 237 nucleotides in length cloned in the coding orientation. Top line, CP-GFP expression was induced for 2 h in 2% galactose medium devoid of methionine. Bottom line, CP-GFP expression was induced for2hin2% glycerol medium devoid of methionine. (C) Reporter RNAs encompassing two subfragments of 100 and 137 nucleotides of the human OXA1 3′ UTR amplified with specific oligonucleotides, and cloned in the unique _Sma_I site of the pIIIA/MS2–2 plasmid (see MATERIALS AND METHODS). The insert orientations inside the plasmid were checked by PCR by using an internal pIIIA/MS2-2 oligonucleotide. The CP-GFP expression was induced for 2 h in 2% glycerol medium devoid of methionine.
Similar articles
- Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae.
Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, Gerst JE. Gadir N, et al. RNA. 2011 Aug;17(8):1551-65. doi: 10.1261/rna.2621111. Epub 2011 Jun 24. RNA. 2011. PMID: 21705432 Free PMC article. - Insertion of proteins into the inner membrane of mitochondria: the role of the Oxa1 complex.
Stuart R. Stuart R. Biochim Biophys Acta. 2002 Sep 2;1592(1):79-87. doi: 10.1016/s0167-4889(02)00266-5. Biochim Biophys Acta. 2002. PMID: 12191770 Review. - In yeast, the 3' untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria.
Corral-Debrinski M, Blugeon C, Jacq C. Corral-Debrinski M, et al. Mol Cell Biol. 2000 Nov;20(21):7881-92. doi: 10.1128/MCB.20.21.7881-7892.2000. Mol Cell Biol. 2000. PMID: 11027259 Free PMC article. - The membrane insertase Oxa1 is required for efficient import of carrier proteins into mitochondria.
Hildenbeutel M, Theis M, Geier M, Haferkamp I, Neuhaus HE, Herrmann JM, Ott M. Hildenbeutel M, et al. J Mol Biol. 2012 Nov 2;423(4):590-9. doi: 10.1016/j.jmb.2012.07.018. Epub 2012 Jul 27. J Mol Biol. 2012. PMID: 22846909 - Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes.
Bonnefoy N, Fiumera HL, Dujardin G, Fox TD. Bonnefoy N, et al. Biochim Biophys Acta. 2009 Jan;1793(1):60-70. doi: 10.1016/j.bbamcr.2008.05.004. Epub 2008 May 15. Biochim Biophys Acta. 2009. PMID: 18522806 Free PMC article. Review.
Cited by
- Direct evidence of CRISPR-Cas9-mediated mitochondrial genome editing.
Bi R, Li Y, Xu M, Zheng Q, Zhang DF, Li X, Ma G, Xiang B, Zhu X, Zhao H, Huang X, Zheng P, Yao YG. Bi R, et al. Innovation (Camb). 2022 Sep 27;3(6):100329. doi: 10.1016/j.xinn.2022.100329. eCollection 2022 Nov 8. Innovation (Camb). 2022. PMID: 36275864 Free PMC article. - MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis.
Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. Kren BT, et al. RNA Biol. 2009 Jan-Mar;6(1):65-72. doi: 10.4161/rna.6.1.7534. Epub 2009 Jan 1. RNA Biol. 2009. PMID: 19106625 Free PMC article. - Biogenesis and dynamics of mitochondria during the cell cycle: significance of 3'UTRs.
Martínez-Diez M, Santamaría G, Ortega AD, Cuezva JM. Martínez-Diez M, et al. PLoS One. 2006 Dec 20;1(1):e107. doi: 10.1371/journal.pone.0000107. PLoS One. 2006. PMID: 17205111 Free PMC article. - High-throughput polyribosome fractionation.
Wang Y, Ringquist S, Cho AH, Rondeau G, Welsh J. Wang Y, et al. Nucleic Acids Res. 2004 Jun 1;32(10):e79. doi: 10.1093/nar/gnh077. Nucleic Acids Res. 2004. PMID: 15173352 Free PMC article. - Codon optimization is an essential parameter for the efficient allotopic expression of mtDNA genes.
Lewis CJ, Dixit B, Batiuk E, Hall CJ, O'Connor MS, Boominathan A. Lewis CJ, et al. Redox Biol. 2020 Feb;30:101429. doi: 10.1016/j.redox.2020.101429. Epub 2020 Jan 11. Redox Biol. 2020. PMID: 31981894 Free PMC article.
References
- Bassell, G.J., Oleynikov, Y., and Singer, R.H. (1999). The travels of mRNAs through all cells large and small. FASEB J. 13, 447–454. - PubMed
- Beach, D.L., Salmon, E.D., and Bloom, K. (1999). Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9, 569–578. - PubMed
- Bonnefoy, N., Kermorgant, M., Groudinsky, O., and Dujardin, G. (2000). The respiratory gene OXA1 has two fission yeast orthologs which together encode a function essential for cellular viability. Mol. Microbiol. 35, 1135–1145. - PubMed
- Bonnefoy, N., Kermorgant, M., Groudinsky, O., Minet, M., Slonimski, P.P., and Dujardin, G. (1994). Cloning of a human gene involved in cytochrome oxidase assembly by functional complementation of an oxa1– mutation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 91, 11978–11982. - PMC - PubMed
- Chartrand, P., Meng, X.-H., Singer, R.H., and Long, R.M. (1999). Structural elements required for the localization of Ash1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9, 333–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases