Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death - PubMed (original) (raw)
Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death
Karin Lykke-Andersen et al. Mol Cell Biol. 2003 Oct.
Abstract
Csn2 (Trip15/Cops2/Alien) encodes the second subunit of the COP9 signalosome (CSN), an eight-subunit heteromeric complex homologous to the lid subcomplex of the 26S proteasome. CSN is a regulator of SCF (Skp1-cullin-F-box protein)ubiquitin ligases, mostly through the enzymatic activity that deconjugates the ubiquitin-like protein Nedd8 from the SCF Cul1 component. In addition, CSN associates with protein kinase activities targeting p53, c-Jun, and IkappaB for phosphorylation. Csn2 also interacts with and regulates a subset of nuclear hormone receptors and is considered a novel corepressor. We report that targeted disruption of Csn2 in mice caused arrest of embryo development at the peri-implantation stage. Csn2(-/-) blastocysts failed to outgrow in culture and exhibited a cell proliferation defect in inner cell mass, accompanied by a slight decrease in Oct4. In addition, lack of Csn2 disrupted the CSN complex and resulted in a drastic increase in cyclin E, supporting a role for CSN in cooperating with the SCF-ubiquitin-proteasome system to regulate protein turnover. Furthermore, Csn2(-/-) embryos contained elevated levels of p53 and p21, which may contribute to premature cell cycle arrest of the mutant.
Figures
FIG. 1.
Targeted disruption of mouse Csn2. (A) A diagram of the targeting construct and the genomic structure of mouse Csn2 locus. The locations of the 5′ and 3′ probes and the relative sizes of the restriction fragments of the wild-type or targeted alleles from the homologous recombination are shown. (B) Identification of homologous recombinants. Genomic DNA from independent ES cell colonies was digested with _Eco_RI and analyzed by Southern blot with the 5′ probe to identify the clones carrying the 3.5-kb fragment specific for targeted allele (+/− lanes). Three out of six colonies shown indicated successful targeting. These three clones were further characterized with the 3′ probe in an _Eco_RV digest to verify the presence of the 13-kb targeting fragment (C).
FIG. 2.
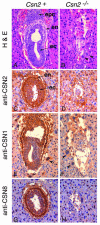
Developmental arrest of Csn2−/− embryos at E6.5. The sagittal sections were stained with hematoxylin and eosin (A and B). The Csn2+ embryos developed highly structured egg cylinder-stage embryos with a central proaminiotic cavity (A), whereas the Csn2−/− embryos had fewer cells and lacked normal architecture and cell differentiation (B). Expression of CSN subunits, as analyzed by immunohistochemical staining with anti-Csn2 (C and D), anti-Csn1 (E and F), or anti-Csn8 (G and H), was present in Csn2+ embryos but absent in Csn2−/− embryos. Sagittal sections (A, B, E, and F) and transverse sections (C, D, G, and H) are shown. Abbreviations: epc, ectoplacental cone; en, extraembryonic endoderm; ec, embryonic ectoderm.
FIG. 3.
Csn2−/− blastocysts failed in outgrowth assay and displayed cell proliferation deficiency. (A) E3.5 blastocysts isolated from Csn2+/− intercrosses were cultured to allow outgrowth. Photographs were taken each day from day 1 to day 5 after isolation. The ICM and the trophoblast cells are marked. ICM development occurred in Csn2+ blastocysts but failed in Csn2−/− blastocysts. (B) Bromodeoxyuridine (BrdU) incorporation during blastocyst outgrowth. Bromodeoxyuridine was added to the medium and incubated for 12 h from 48 h to 60 h (day 2), from 84 h to 96 h (days 3 to 4), or from 108 h to 120 h (days 4 to 5). Blastocysts were fixed at the end of the 12-h labeling period and immunostained with antibromodeoxyuridine antibody. The corresponding nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (DNA). Bromodeoxyuridine incorporation ceased by day 4 in Csn2−/− blastocyst cultures but continued in Csn2+ blastocysts. (C) PCR genotyping of blastocysts showing the three different genotypes: Csn2+/+ , Csn2+/− and Csn2−/−. (D) Expression of Oct4 and Fgf4 in Csn2−/− blastocysts. Hatched blastocysts after 1 day in culture were used for immunofluorescence staining of Oct4 (a and b). Oct4 expression appeared slightly weaker in the mutant. Blastocysts grown for 3 days were subjected to immunofluorescence staining with Fgf4 (c and d). The mutant appeared to have reduced Fgf4 staining in the ICM region but slightly higher staining in the trophoblasts. DAPI staining shows the nuclei DNA of the corresponding blastocysts (e and f).
FIG. 4.
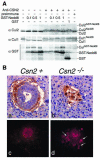
Inactivation of Csn2 leads to Cul1 and Cul2 deneddylation defect and elevated accumulation of cyclin E. (A) HeLa cell extract was supplied with an ATP regeneration system and additional components as indicated. These included polyclonal antiserum against Csn2 (1 μl) or preimmune serum (1 μl); GST-Nedd8, 0.1, 0.5 or 1.0 μg; and GST, 1 μg. The samples were then analyzed by immunoblot with antibodies against Cul1, Cul2, and GST. The anti-Csn2 serum inhibited deneddylation, allowing neddylated cullins to accumulate as driven by active neddylation reactions in the system. (B) Abnormal accumulation of cyclin E in Csn2−/− embryos and blastocysts. Immunohistochemical analysis of E6.5 transverse sections display ubiquitous and high-intensity cyclin E staining in Csn2−/− embryos (b) compared to the weak endoderm staining in normal embryos (a). Immunofluorescence analysis of blastocyst outgrowth at day 2 showed higher cyclin E immunoreaction in the mutant (d) compared to normal blastocysts (c), particularly in trophectoderm cells (d, arrows).
FIG. 5.
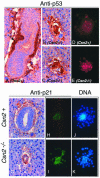
Csn2−/− embryos accumulated elevated levels of p53 and p21. The E6.5 embryos were analyzed by immunohistochemical staining for p53 (A to E), or p21 (F to I). (A) Sagittal section of normal (Csn2+) embryos stained for p53. The arrow points to the extraembryonic endoderm cells that contain p53 staining. (B and C) Csn2−/− mutants contained intense p53 staining in almost all cells of the embryos. (D and E) Blastocysts grown in culture for 2.5 days were used for anti-p53 immunofluorescence staining. The mutant cells (E) contain higher p53 immunoreactions compared to the Csn2+ blastocyst outgrowth (D). Immunohistochemical analysis of p21 on a transverse section of normal embryo (F) compared with Csn2−/− embryos (G) showed more widespread and more intense staining in the mutant. Immunofluorescence analysis of p21 expression on 3-day blastocyst outgrowth (H and I) further confirmed that p21 expression was increased in both presumed ICM and trophectoderm cells of the Csn2−/− mutants. Blastocysts were double stained with DAPI to show the nuclear DNA (J and K).
Similar articles
- Roles of COP9 signalosome in cancer.
Lee MH, Zhao R, Phan L, Yeung SC. Lee MH, et al. Cell Cycle. 2011 Sep 15;10(18):3057-66. doi: 10.4161/cc.10.18.17320. Epub 2011 Sep 15. Cell Cycle. 2011. PMID: 21876386 Free PMC article. Review. - Multiple functions of Jab1 are required for early embryonic development and growth potential in mice.
Tomoda K, Yoneda-Kato N, Fukumoto A, Yamanaka S, Kato JY. Tomoda K, et al. J Biol Chem. 2004 Oct 8;279(41):43013-8. doi: 10.1074/jbc.M406559200. Epub 2004 Aug 6. J Biol Chem. 2004. PMID: 15299027 - The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1.
Yang X, Menon S, Lykke-Andersen K, Tsuge T, Di Xiao, Wang X, Rodriguez-Suarez RJ, Zhang H, Wei N. Yang X, et al. Curr Biol. 2002 Apr 16;12(8):667-72. doi: 10.1016/s0960-9822(02)00791-1. Curr Biol. 2002. PMID: 11967155 - Analysis of the role of COP9 Signalosome (CSN) subunits in K562; the first link between CSN and autophagy.
Pearce C, Hayden RE, Bunce CM, Khanim FL. Pearce C, et al. BMC Cell Biol. 2009 Apr 28;10:31. doi: 10.1186/1471-2121-10-31. BMC Cell Biol. 2009. PMID: 19400951 Free PMC article. - The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis.
Bech-Otschir D, Seeger M, Dubiel W. Bech-Otschir D, et al. J Cell Sci. 2002 Feb 1;115(Pt 3):467-73. doi: 10.1242/jcs.115.3.467. J Cell Sci. 2002. PMID: 11861754 Review.
Cited by
- IP6-assisted CSN-COP1 competition regulates a CRL4-ETV5 proteolytic checkpoint to safeguard glucose-induced insulin secretion.
Lin H, Yan Y, Luo Y, So WY, Wei X, Zhang X, Yang X, Zhang J, Su Y, Yang X, Zhang B, Zhang K, Jiang N, Chow BKC, Han W, Wang F, Rao F. Lin H, et al. Nat Commun. 2021 Apr 28;12(1):2461. doi: 10.1038/s41467-021-22941-3. Nat Commun. 2021. PMID: 33911083 Free PMC article. - The COP9 signalosome and vascular function: intriguing possibilities?
Martin DS, Wang X. Martin DS, et al. Am J Cardiovasc Dis. 2015 Mar 20;5(1):33-52. eCollection 2015. Am J Cardiovasc Dis. 2015. PMID: 26064791 Free PMC article. Review. - The ubiquitin-proteasome system.
Nandi D, Tahiliani P, Kumar A, Chandu D. Nandi D, et al. J Biosci. 2006 Mar;31(1):137-55. doi: 10.1007/BF02705243. J Biosci. 2006. PMID: 16595883 Review. - Roles of COP9 signalosome in cancer.
Lee MH, Zhao R, Phan L, Yeung SC. Lee MH, et al. Cell Cycle. 2011 Sep 15;10(18):3057-66. doi: 10.4161/cc.10.18.17320. Epub 2011 Sep 15. Cell Cycle. 2011. PMID: 21876386 Free PMC article. Review. - Role of individual subunits of the Neurospora crassa CSN complex in regulation of deneddylation and stability of cullin proteins.
Wang J, Hu Q, Chen H, Zhou Z, Li W, Wang Y, Li S, He Q. Wang J, et al. PLoS Genet. 2010 Dec 2;6(12):e1001232. doi: 10.1371/journal.pgen.1001232. PLoS Genet. 2010. PMID: 21151958 Free PMC article.
References
- Akiyama, H., A. Sugiyama, K. Uzawa, N. Fujisawa, Y. Tashiro, and F. Tashiro. 2003. Implication of Trip15/CSN2 in early stage of neuronal differentiation of P19 embryonal carcinoma cells. Brain Res. Dev. Brain Res. 140:45-56. - PubMed
- Altincicek, B., S. P. Tenbaum, U. Dressel, D. Thormeyer, R. Renkawitz, and A. Baniahmad. 2000. Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J. Biol. Chem. 275:7662-7667. - PubMed
- Cohen, H., A. Azriel, T. Cohen, D. Meraro, S. Hashmueli, D. Bech-Otschir, R. Kraft, W. Dubiel, and B. Z. Levi. 2000. Interaction between interferon consensus sequence-binding protein and COP9/signalosome subunit CSN2 (Trip15). A possible link between interferon regulatory factor signaling and the COP9/signalosome. J. Biol. Chem. 275:39081-39089. - PubMed
- Cope, G. A., G. S. Suh, L. Aravind, S. E. Schwarz, S. L. Zipursky, E. V. Koonin, and R. J. Deshaies. 2002. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298:608-611. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL64641/HL/NHLBI NIH HHS/United States
- ES11384/ES/NIEHS NIH HHS/United States
- U19 ES011384/ES/NIEHS NIH HHS/United States
- GM61812/GM/NIGMS NIH HHS/United States
- R01 HL064641/HL/NHLBI NIH HHS/United States
- AR49496/AR/NIAMS NIH HHS/United States
- R01 AR049496/AR/NIAMS NIH HHS/United States
- R01 GM061812/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous