Evaluation of aldehyde dehydrogenase 1 promoter polymorphisms identified in human populations - PubMed (original) (raw)
Comparative Study
Evaluation of aldehyde dehydrogenase 1 promoter polymorphisms identified in human populations
John P Spence et al. Alcohol Clin Exp Res. 2003 Sep.
Abstract
Background: Cytosolic aldehyde dehydrogenase, or ALDH1A1, functions in ethanol detoxification, metabolism of neurotransmitters, and synthesis of retinoic acid. Because the promoter region of a gene can influence gene expression, the ALDH1A1 promoter regions were studied to identify polymorphism, to assess their functional significance, and to determine whether they were associated with a risk for developing alcoholism.
Methods: Sequence analysis was performed in the promoter region by using Asian, Caucasian, and African American subjects. The resulting polymorphisms were assessed for frequency in Asian, Caucasian, Jewish, and African American populations and tested for associations with alcohol dependence in Asian and African American populations of alcoholics and controls. The functional significance of each polymorphism was determined through in vitro expression analysis by using HeLa and HepG2 cells.
Results: Two polymorphisms, a 17 base pair (bp) deletion (-416/-432) and a 3 bp insertion (-524), were discovered in the ALDH1A1 promoter region: ALDH1A1*2 and ALDH1A1*3, respectively. ALDH1A1*2 was observed at frequencies of 0.035, 0.023, 0.023, and 0.012 in the Asian, Caucasian, Jewish, and African American populations, respectively. ALDH1A1*3 was observed only in the African American population, at a frequency of 0.029. By using HeLa and HepG2 cells for in vitro expression, the activity of the luciferase reporter gene was significantly decreased after transient transfection of ALDH1A1*3-luciferase compared with the wild-type construct ALDH1A1*1-luciferase. In an African American population, a trend for higher frequencies of the ALDH1A1*2 and ALDH1A1*3 alleles was observed in a population of alcoholics (p = 0.03 and f = 0.12, respectively) compared with the control population.
Conclusions: ALDH1A1*2 and ALDH1A1*3 may influence ALDH1A1 gene expression. Both ALDH1A1*2 and ALDH1A1*3 produce a trend in an African American population that may be indicative of an association with alcoholism; however, more samples are required to validate this observation. The underlying mechanisms contributing to these trends are still unknown.
Figures
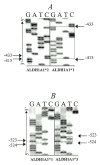
Fig. 1
Sequence analysis of polymorphism in the human ALDH1A1 promoter region. (A) ALDH1A1*2: the box highlights the 17-bp sequence (−416/−432) that was deleted from ALDH1A1*1 to generate ALDH1A1*2. (B) ALDH1A1*3: the box designates the 3-bp insertion (−524). Arrows indicate common flanking nucleotides, and G, A, T, and C represent guanine, adenine, thiamine, and cytosine, respectively.
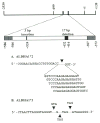
Fig. 2
The human ALDH1A1 promoter region. Sequence analysis was performed on the region spanning −699 to +139 with respect to the transcriptional start point (+1). The magnified region extending from −586 to −372 designates the fragment that was amplified during the genotyping assay. Both polymorphisms are distinguished in this diagram: the 3-bp insertion (−524) and the 17-bp deletion (−416/−432). (A) The deletion of five possible fragments results in the same ALDH1A1*2 sequence due to flanking 5′-GGTC-3′ repeats. (B) Three possible insertions at −524, −523, or −519 generate the same ALDH1A1*3 sequence.
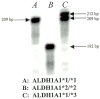
Fig. 3
Autoradiogram of ALDH1A1*1, ALDH1A1*2, and ALDH1A1*3 genotypes. PCR was used to generate [_α_-33P]deoxycytidine triphosphate-radiolabeled fragments that were later separated on a 6% acrylamide gel by using electrophoresis. Each allele, ALDH1A1*1, ALDH1A1*2, and ALDH1A1*3, can be readily identified with this procedure. (A) A homozygous Caucasian with the genotype ALDH1A1*1/*1, the wild-type genotype. (B) A homozygous Asian with the genotype ALDHlA1*2/*2. (C) A heterozygous African American with the genotype ALDH1A1*1/*3.
Fig. 4
Functional importance of ALDH1A1 promoter polymorphisms. Plasmids ALDH1A1*1, ALDH1A1*2, and ALDH1A1*3 were transfected into HepG2 and HeLa cells; luc+ denotes the luciferase gene. Each respective fragment amplified from the ALDH1A1 promoter extended from −659 to +39 with respect to the transcriptional start point, and the polymorphisms, the deletion (−416/−432) and the insertion (−524), are noted. The activity of each construct was normalized by co-transfection of the internal control plasmid, pRL-CMV, and was expressed as fold change compared with the activity of ALDH1A1*1-luc. The mean ± SEM are representative of the results from three to six independent transfections performed in triplicate with two different plasmid preparations. The significance of the difference between mean values within and between multiple constructs was analyzed with ANOVA. ALDH1A1*3-luc significantly decreased luciferase expression in both Hep G2 and HeLa cells compared with ALDH1A1*1-luc (*p < 0.05). No significant change in expression was associated with ALDH1A1*2-luc.
Similar articles
- Association of ALDH1 promoter polymorphisms with alcohol-related phenotypes in Trinidad and Tobago.
Moore S, Montane-Jaime K, Shafe S, Joseph R, Crooks H, Carr LG, Ehlers CL. Moore S, et al. J Stud Alcohol Drugs. 2007 Mar;68(2):192-6. doi: 10.15288/jsad.2007.68.192. J Stud Alcohol Drugs. 2007. PMID: 17286337 - Association of ALDH1 promoter polymorphisms with alcohol-related phenotypes in southwest California Indians.
Ehlers CL, Spence JP, Wall TL, Gilder DA, Carr LG. Ehlers CL, et al. Alcohol Clin Exp Res. 2004 Oct;28(10):1481-6. doi: 10.1097/01.alc.0000141821.06062.20. Alcohol Clin Exp Res. 2004. PMID: 15597079 - Haplotype-based study of the association of alcohol-metabolizing genes with alcohol dependence in four independent populations.
Liu J, Zhou Z, Hodgkinson CA, Yuan Q, Shen PH, Mulligan CJ, Wang A, Gray RR, Roy A, Virkkunen M, Goldman D, Enoch MA. Liu J, et al. Alcohol Clin Exp Res. 2011 Feb;35(2):304-16. doi: 10.1111/j.1530-0277.2010.01346.x. Epub 2010 Nov 17. Alcohol Clin Exp Res. 2011. PMID: 21083667 Free PMC article. - Variations in alcohol-metabolizing enzymes in people of East Indian and African descent from Trinidad and Tobago.
Moore S, Montane-Jaime LK, Carr LG, Ehlers CL. Moore S, et al. Alcohol Res Health. 2007;30(1):28-30. Alcohol Res Health. 2007. PMID: 17718398 Free PMC article. Review. - Health-related effects of genetic variations of alcohol-metabolizing enzymes in African Americans.
Scott DM, Taylor RE. Scott DM, et al. Alcohol Res Health. 2007;30(1):18-21. Alcohol Res Health. 2007. PMID: 17718396 Free PMC article. Review.
Cited by
- Role of Genetic Polymorphisms in Drug-Metabolizing Enzyme-Mediated Toxicity and Pharmacokinetic Resistance to Anti-Cancer Agents: A Review on the Pharmacogenomics Aspect.
Narendra G, Choudhary S, Raju B, Verma H, Silakari O. Narendra G, et al. Clin Pharmacokinet. 2022 Nov;61(11):1495-1517. doi: 10.1007/s40262-022-01174-7. Epub 2022 Sep 30. Clin Pharmacokinet. 2022. PMID: 36180817 Review. - Disulfiram suppressed ethanol promoted RANKL-induced osteoclastogenesis in vitro and ethanol-induced osteoporosis in vivo via ALDH1A1-NFATc1 axis.
Jia Y, Jiang J, Zhao K, Zhang T, Sun P, Peng J, Yang Q, Qian Y. Jia Y, et al. Aging (Albany NY). 2019 Oct 8;11(19):8103-8119. doi: 10.18632/aging.102279. Epub 2019 Oct 8. Aging (Albany NY). 2019. PMID: 31596733 Free PMC article. - Species and inter-individual differences in metabolic capacity of di(2-ethylhexyl)phthalate (DEHP) between human and mouse livers.
Ito Y, Kamijima M, Hasegawa C, Tagawa M, Kawai T, Miyake M, Hayashi Y, Naito H, Nakajima T. Ito Y, et al. Environ Health Prev Med. 2014 Mar;19(2):117-25. doi: 10.1007/s12199-013-0362-6. Epub 2013 Sep 28. Environ Health Prev Med. 2014. PMID: 24078404 Free PMC article. - The role of aldehyde dehydrogenase-1 (ALDH1A1) polymorphisms in harmful alcohol consumption in a Finnish population.
Lind PA, Eriksson CJ, Wilhelmsen KC. Lind PA, et al. Hum Genomics. 2008 Sep;3(1):24-35. doi: 10.1186/1479-7364-3-1-24. Hum Genomics. 2008. PMID: 19129088 Free PMC article. - Association of ADH and ALDH genes with alcohol dependence in the Irish Affected Sib Pair Study of alcohol dependence (IASPSAD) sample.
Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van den Oord EJ, Alexander J, Jiang C, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Kuo PH, et al. Alcohol Clin Exp Res. 2008 May;32(5):785-95. doi: 10.1111/j.1530-0277.2008.00642.x. Epub 2008 Mar 4. Alcohol Clin Exp Res. 2008. PMID: 18331377 Free PMC article.
References
- Ambroziak W, Izaguirre G, Pietruszko R. Metabolism of retinaldehyde and other aldehydes in soluble extracts of human liver and kidney. J Biol Chem. 1999;274:33366–33373. - PubMed
- Brien JF, Loomis CW. Pharmacology of acetaldehyde. Can J Physiol Pharmacol. 1983;61:1–22. - PubMed
- Carr LG, Foroud T, Stewart T, Castelluccio P, Edenberg HJ, Li T-K. Influence of ADH1B polymorphism on alcohol use and its subjective effects in a Jewish population. Am J Med. 2002;112:138–143. - PubMed
- Chan AW. Racial differences in alcohol sensitivity. Alcohol Alcohol. 1986;21:93–104. - PubMed
- Crabb DW, Pinairs J, Hasanadka R, Fang M, Leo MA, Lieber CS, Tsukamoto H, Motomura K, Miyahara T, Ohata M, Bosron W, Sanghani S, Kedishvili N, Shiraishi H, Yokoyama H, Miyagi M, Ishii H, Bergheim I, Menzl I, Parlesak A, Bode C. Alcohol and retinoids. Alcohol Clin Exp Res. 2001;25(Suppl 5):207S–217S. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AA05546/AA/NIAAA NIH HHS/United States
- AA11257/AA/NIAAA NIH HHS/United States
- R29 AA011257/AA/NIAAA NIH HHS/United States
- U24-AA-11898/AA/NIAAA NIH HHS/United States
- AA00269/AA/NIAAA NIH HHS/United States
- P60 AA007611/AA/NIAAA NIH HHS/United States
- F31 AA005546/AA/NIAAA NIH HHS/United States
- P50 AA007611/AA/NIAAA NIH HHS/United States
- R01 AA011257/AA/NIAAA NIH HHS/United States
- AA07611/AA/NIAAA NIH HHS/United States
- K02 AA000269/AA/NIAAA NIH HHS/United States
- R01 AA010707/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous