Lsh, a modulator of CpG methylation, is crucial for normal histone methylation - PubMed (original) (raw)
Lsh, a modulator of CpG methylation, is crucial for normal histone methylation
Qingsheng Yan et al. EMBO J. 2003.
Abstract
Methylation of histone tails and CpG methylation are involved in determining heterochromatin structure, but their cause and effect relationship has not been resolved as yet in mammals. Here we report that Lsh, a member of the SNF2 chromatin remodeling family, controls both types of epigenetic modifications. Lsh has been shown to be associated with pericentromeric heterochromatin and to be required for normal CpG methylation at pericentromeric sequences. Loss of Lsh, in Lsh-deficient mice, results in accumulation of di- and tri-methylated histone 3 at lysine 4 (H3-K4me) at pericentromeric DNA and other repetitive sequences. In contrast, di- or tri-methylation of H3-K9 and distribution of HP1 appear unchanged after Lsh deletion, suggesting independent regulatory mechanisms for H3-K4 or K9 methylation. Experimental DNA demethylation with 5'-azacytidine results in a similar increase of H3-K4me. These results support the model that loss of CpG methylation caused by Lsh deficiency antecedes elevation of H3-K4me. Thus, Lsh is crucial for the formation of normal heterochromatin, implying a functional role for Lsh in the regulation of transcription and mitosis.
Figures
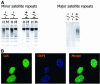
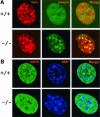
Fig. 1. Lsh associates with pericentromeric heterochromatin. (A) Lsh controls CpG methylation at pericentromeric regions in mice. Southern analysis of minor and major satellite sequences utilizing the CpG-sensitive restriction enzyme _Hpa_II (H) and the methylation-insensitive restriction enzymes _Msp_I (M) (for minor) or _Mae_II (for major) comparing Lsh+/+ and Lsh–/– MEFs. (B) Lsh localizes to pericentromeric heterochromatin. Immunofluorescence staining of Flag-tagged Lsh after induction in 3T3 fibroblasts utilizing an anti-Flag antibody. Nuclei are stained with DAPI.
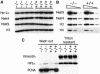
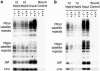
Fig. 2. Lsh does not regulate global histone methylation levels. (A) Loss of Lsh does not alter H3-K9me, H3-K4me or Hp1 protein levels. Western analysis of nuclear extracts derived from four Lsh-deficient embryos (gestation day 14) and their wild-type littermates. (B) Titration of nuclear extracts of one wild-type and one Lsh-deficient embryo. (C) Loss of Lsh does not alter Hp1 binding to chromatin. Lsh-deficient and Lsh wild-type MEFs are compared for resistance to Triton X-100 washes followed by western analysis to detect HP1 or PCNA and Vimentin as control.
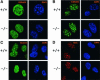
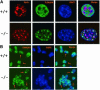
Fig. 3. Loss of Lsh does not alter H3-K9 methylation or HP1 association at pericentromeric chromatin. (A) Lsh-deficient and wild-type MEFs are immunostained for detection of dimethylated H3-K9 and counterstained with DAPI. (B) The same as in (A) but antiserum raised against trimethylated H3-K9 peptide was used. (C) The same as in (A) but antiserum raised against the branched dimethylated H3-K9 peptide was used. (D) Lsh-deficient and wild-type MEFs are immunostained for detection of HP1 and counterstained with DAPI.
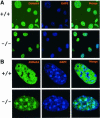
Fig. 4. Absence of Lsh leads to accumulation of dimethylated H3-K4 at pericentromeric heterochromatin. (A) Lsh-deficient and wild-type MEFs are immunostained for detection of dimethylated H3-K4 and counterstained with DAPI. (B) The same as in (A), but at higher magnification.
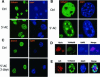
Fig. 5. Co-localization of methylated H3-K4 with HP1. (A) Lsh-deficient and wild-type MEFs are double stained for detection of dimethylated H3-K4 and HP1. (B) Lsh-deficient and wild-type MEFs are immunostained for detection of acetylated H4-K16 and counterstained with DAPI.
Fig. 6. Lsh deficiency raises trimethylated H3-K4 at pericentromeric regions. (A) Lsh-deficient and wild-type MEFs are double stained for detection of trimethylated H3-K4, HP1 and counterstained with Dapi. (B) Lsh-deficient and wild-type MEFs are treated with colcemid and the metaphase spread immunostained for detection of dimethylated H3-K4 and counterstained with DAPI.
Fig. 7. Loss of Lsh increases H3-K4 methylation at repetitive sequences. (A) Chromatin immunoprecipitation with specific antibodies raised against dimethylated H3-K4 and dimethylated H3-K9 were performed using nuclear extracts derived from Lsh-deficient and wild-type MEFs, followed by PCR analysis for detection of minor and major satellite sequences, the IAP and Line elements. (B) ChIPs was performed as in (A) with specific antibodies raised against trimethylated H3-K4 and trimethylated H3-K9.
Fig. 8. Derepression of IAP transcripts in the absence of Lsh. (A) Diagram of the regions for single copy genes that have been previously examined by Southern analysis and found to be hypomethylated (H19, globin, pgk-2) or unaltered (Igf2R, ARPT). Triangles indicate the position of the primers used in ChIps experiments located at methylation-sensitive _Hha_I sites. (B) Chromatin immunoprecipitation with specific antibodies raised against dimethylated H3-K4 and dimethylated H3-K9 were performed using nuclear extracts derived from Lsh-deficient and wild-type MEFs followed by PCR analysis for detection of the indicated single copy genes. (C) RT–PCR analysis of cDNA derived from Lsh-deficient and wild-type MEFs for detection of IAP and Line1 transcripts with actin as a control. For better comparison cDNA was titrated in dilution steps of 1:3. Amplification in the absence of reverse transcriptase served as control. (D) 5′-azacytidine treatment reduces CpG methylation at pericentromeric DNA. Southern analysis of minor satellite sequences utilizing the CpG sensitive restriction enzyme _Hpa_II (H) and the methylation-insensitive restriction enzyme _Msp_I (M) comparing untreated 3T3 fibroblasts and cells treated for 5 days with 5′-azacytidine. (E) 5′-azacytidine does not alter global H3 methylation levels. Western analysis of nuclear extracts derived from untreated and 5′-azacytidine-treated 3T3 fibroblasts for detection of methylated H3 and Hp1.
Fig. 9. Global demethylation causes redistribution of methylated H3-K4 at pericentromeric heterochromatin. (A and B) Demethylation by 5′-azacytidine increases H3-K4 methylation. 3T3 fibroblasts were treated for 3 days with 5′-azacytidine and immunostained for detection of dimethylated H3-K4 and counterstained with DAPI. (C) Same as in (A), but antiserum raised against the branched dimethylated H3-K9 peptide was used. (D) Prolonged treatment of 3T3 fibroblasts (for 5 days) with 5′-azacytidine followed by immunostaining for detection of HP1, trimethylated H3-K9 and counterstain with DAPI. (E) Same as in (D) but Flag-tagged Lsh was induced over the last 24 h and cells were then immunostained using antiserum for detection of the Flag-tag Lsh protein or detection of trimethylated H3-K4.
Similar articles
- Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin.
Yan Q, Cho E, Lockett S, Muegge K. Yan Q, et al. Mol Cell Biol. 2003 Dec;23(23):8416-28. doi: 10.1128/MCB.23.23.8416-8428.2003. Mol Cell Biol. 2003. PMID: 14612388 Free PMC article. - Cell cycle-dependent accumulation of histone H3.3 and euchromatic histone modifications in pericentromeric heterochromatin in response to a decrease in DNA methylation levels.
Sugimura K, Fukushima Y, Ishida M, Ito S, Nakamura M, Mori Y, Okumura K. Sugimura K, et al. Exp Cell Res. 2010 Oct 15;316(17):2731-46. doi: 10.1016/j.yexcr.2010.06.016. Epub 2010 Jun 25. Exp Cell Res. 2010. PMID: 20599948 - Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin.
Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Lehnertz B, et al. Curr Biol. 2003 Jul 15;13(14):1192-200. doi: 10.1016/s0960-9822(03)00432-9. Curr Biol. 2003. PMID: 12867029 - Lsh, a guardian of heterochromatin at repeat elements.
Muegge K. Muegge K. Biochem Cell Biol. 2005 Aug;83(4):548-54. doi: 10.1139/o05-119. Biochem Cell Biol. 2005. PMID: 16094458 Review.
Cited by
- Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis.
Mathieu O, Probst AV, Paszkowski J. Mathieu O, et al. EMBO J. 2005 Aug 3;24(15):2783-91. doi: 10.1038/sj.emboj.7600743. Epub 2005 Jul 7. EMBO J. 2005. PMID: 16001083 Free PMC article. - Treatment of breast cancer cells with DNA demethylating agents leads to a release of Pol II stalling at genes with DNA-hypermethylated regions upstream of TSS.
Tao Y, Liu S, Briones V, Geiman TM, Muegge K. Tao Y, et al. Nucleic Acids Res. 2011 Dec;39(22):9508-20. doi: 10.1093/nar/gkr611. Epub 2011 Aug 31. Nucleic Acids Res. 2011. PMID: 21880597 Free PMC article. - A role for LSH in facilitating DNA methylation by DNMT1 through enhancing UHRF1 chromatin association.
Han M, Li J, Cao Y, Huang Y, Li W, Zhu H, Zhao Q, Han JJ, Wu Q, Li J, Feng J, Wong J. Han M, et al. Nucleic Acids Res. 2020 Dec 2;48(21):12116-12134. doi: 10.1093/nar/gkaa1003. Nucleic Acids Res. 2020. PMID: 33170271 Free PMC article. - Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline.
Crichton JH, Dunican DS, Maclennan M, Meehan RR, Adams IR. Crichton JH, et al. Cell Mol Life Sci. 2014 May;71(9):1581-605. doi: 10.1007/s00018-013-1468-0. Epub 2013 Sep 18. Cell Mol Life Sci. 2014. PMID: 24045705 Free PMC article. Review. - The ATP binding site of the chromatin remodeling homolog Lsh is required for nucleosome density and de novo DNA methylation at repeat sequences.
Ren J, Briones V, Barbour S, Yu W, Han Y, Terashima M, Muegge K. Ren J, et al. Nucleic Acids Res. 2015 Feb 18;43(3):1444-55. doi: 10.1093/nar/gku1371. Epub 2015 Jan 10. Nucleic Acids Res. 2015. PMID: 25578963 Free PMC article.
References
- Ahmad K. and Henikoff,S. (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell, 9, 1191–1200. - PubMed
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. - PubMed
- Becker P.B. and Horz,W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., 71, 247–273. - PubMed
- Bernard P., Maure,J.F., Partridge,J.F., Genier,S., Javerzat,J.P. and Allshire,R.C. (2001) Requirement of heterochromatin for cohesion at centromeres. Science, 294, 2539–2542. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases