Identifying biological themes within lists of genes with EASE - PubMed (original) (raw)
Identifying biological themes within lists of genes with EASE
Douglas A Hosack et al. Genome Biol. 2003.
Abstract
EASE is a customizable software application for rapid biological interpretation of gene lists that result from the analysis of microarray, proteomics, SAGE and other high-throughput genomic data. The biological themes returned by EASE recapitulate manually determined themes in previously published gene lists and are robust to varying methods of normalization, intensity calculation and statistical selection of genes. EASE is a powerful tool for rapidly converting the results of functional genomics studies from 'genes' to 'themes'.
Figures
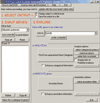
Figure 1
The EASE user interface is designed for quick annotation and analysis of gene lists. Gene identifiers are pasted into the 'INPUT GENES' section, and the processes of linking to online tools, over-representation analysis or annotation are launched with buttons in the 'EXPLORE' section. Annotation data can be automatically retrieved from the internet and stored into local data files by clicking the 'update with the most recent online data' button.
Similar articles
- Extracting biological meaning from large gene lists with DAVID.
Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Huang da W, et al. Curr Protoc Bioinformatics. 2009 Sep;Chapter 13:Unit 13.11. doi: 10.1002/0471250953.bi1311s27. Curr Protoc Bioinformatics. 2009. PMID: 19728287 - MILANO--custom annotation of microarray results using automatic literature searches.
Rubinstein R, Simon I. Rubinstein R, et al. BMC Bioinformatics. 2005 Jan 20;6:12. doi: 10.1186/1471-2105-6-12. BMC Bioinformatics. 2005. PMID: 15661078 Free PMC article. - PubMatrix: a tool for multiplex literature mining.
Becker KG, Hosack DA, Dennis G Jr, Lempicki RA, Bright TJ, Cheadle C, Engel J. Becker KG, et al. BMC Bioinformatics. 2003 Dec 10;4:61. doi: 10.1186/1471-2105-4-61. BMC Bioinformatics. 2003. PMID: 14667255 Free PMC article. - List of lists-annotated (LOLA): a database for annotation and comparison of published microarray gene lists.
Cahan P, Ahmad AM, Burke H, Fu S, Lai Y, Florea L, Dharker N, Kobrinski T, Kale P, McCaffrey TA. Cahan P, et al. Gene. 2005 Oct 24;360(1):78-82. doi: 10.1016/j.gene.2005.07.008. Epub 2005 Sep 2. Gene. 2005. PMID: 16140476 Review. - Where are we in genomics?
Hocquette JF. Hocquette JF. J Physiol Pharmacol. 2005 Jun;56 Suppl 3:37-70. J Physiol Pharmacol. 2005. PMID: 16077195 Review.
Cited by
- Proteomic Profiling of Endothelial Cell Secretomes After Exposure to Calciprotein Particles Reveals Downregulation of Basement Membrane Assembly and Increased Release of Soluble CD59.
Stepanov A, Shishkova D, Markova V, Markova Y, Frolov A, Lazebnaya A, Oshchepkova K, Perepletchikova D, Smirnova D, Basovich L, Repkin E, Kutikhin A. Stepanov A, et al. Int J Mol Sci. 2024 Oct 23;25(21):11382. doi: 10.3390/ijms252111382. Int J Mol Sci. 2024. PMID: 39518935 Free PMC article. - Circulating proteins linked to apoptosis processes and fast development of end-stage kidney disease in diabetes.
Ihara K, Satake E, Wilson PC, Krolewski B, Kobayashi H, Md Dom ZI, Ricca J, Wilson J, Dreyfuss JM, Niewczas MA, Doria A, Nelson RG, Pezzolesi MG, Humphreys BD, Duffin K, Krolewski AS. Ihara K, et al. JCI Insight. 2024 Oct 22;9(20):e178373. doi: 10.1172/jci.insight.178373. JCI Insight. 2024. PMID: 39435665 Free PMC article. - Translation of genome-wide association study: from genomic signals to biological insights.
Bruner WS, Grant SFA. Bruner WS, et al. Front Genet. 2024 Oct 3;15:1375481. doi: 10.3389/fgene.2024.1375481. eCollection 2024. Front Genet. 2024. PMID: 39421299 Free PMC article. Review. - DAVID Ortholog: an integrative tool to enhance functional analysis through orthologs.
Sherman BT, Panzade G, Imamichi T, Chang W. Sherman BT, et al. Bioinformatics. 2024 Oct 1;40(10):btae615. doi: 10.1093/bioinformatics/btae615. Bioinformatics. 2024. PMID: 39412445 Free PMC article. - Current approaches and outstanding challenges of functional annotation of metabolites: a comprehensive review.
Nguyen QH, Nguyen H, Oh EC, Nguyen T. Nguyen QH, et al. Brief Bioinform. 2024 Sep 23;25(6):bbae498. doi: 10.1093/bib/bbae498. Brief Bioinform. 2024. PMID: 39397425 Free PMC article. Review.
References
- Heller MJ. DNA microarray technology: devices, systems, and applications. Annu Rev Biomed Eng. 2002;4:129–153. - PubMed
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32 Suppl:496–501. - PubMed
- Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet. 2002;32 Suppl:502–508. - PubMed
- Pruitt KD, Katz KS, Sicotte H, Maglott DR. Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet. 2000;16:44–47. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical