Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo - PubMed (original) (raw)
Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo
Maarten Hoek et al. Proc Natl Acad Sci U S A. 2003.
Abstract
De novo chromatin assembly maintains histone density on the daughter strands in the wake of the replication fork. The heterotrimer chromatin assembly factor 1 (CAF-1) couples DNA replication to histone deposition in vitro, but is not essential for yeast cell proliferation. Depletion of CAF-1 in human cell lines demonstrated that CAF-1 was required for efficient progression through S-phase. Cells lacking CAF-1 accumulated in early and mid S-phase and replicated DNA slowly. The checkpoint kinase Chk1, but not Chk2, was phosphorylated in response to CAF-1 depletion, consistent with a DNA replication defect. CAF-1-depleted cell extracts completely lacked DNA replication-coupled chromatin assembly activity, suggesting that CAF-1 is required for efficient S-phase progression in human cells. These results indicate that, in contrast to yeast, human CAF-1 is necessary for coupling chromatin assembly with DNA replication.
Figures
Fig. 1.
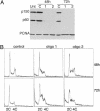
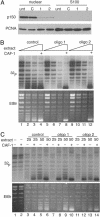
RNAi of CAF-1 induces the accumulation of cells in S-phase (A) Western blot analysis of total cell extract of cells treated with control oligo or with two different oligos targeting CAF-1 p150. Equal amounts of cell extract were loaded and probed with antibodies against CAF-1 p150, CAF-1 p60, and PCNA. Note that the bulk of p60 is depleted in p150 knockdown cells. (B) Cell cycle analysis of control and CAF-1-depleted cells at 48 and 72 h after transfection. DNA content was measured by flow cytometry.
Fig. 2.
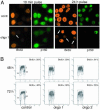
Loss of CAF-1 leads to slow S-phase progression. (A) CAF-1 knockdown cells pulsed for 10 min (48 h after transfection) or 24 h (36–60 h after transfection) with 40 μM BrdUrd and stained with antibodies specific for BrdUrd and p150. Control cells display high levels of BrdUrd incorporation, whereas CAF-1-depleted cells do not incorporate BrdUrd efficiently (arrows). (B) Two-dimensional flow cytometry analysis monitoring BrdUrd incorporation and DNA content. Cells were pulsed with BrdUrd for 10 min, fixed, and stained with FITC-conjugated anti-BrdUrd antibody and propidium iodide to stain the DNA. BrdUrd intensity is plotted on a log scale on the y axis, and DNA content is plotted on the x axis. G1, G2, and S-phase populations are gated in each sample, and the numbers in the upper right indicate the percentage of BrdUrd-positive cells in each sample.
Fig. 3.
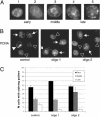
CAF-1-depleted cells assemble normal replication foci and accumulate in early to mid S-phase. (A) Cells were stained for chromatin-bound PCNA and scored for replication patterns. Examples of early, mid, and late S-phase patterns are provided. (B) Late S-phase patterns are easily seen in control cells but are found less frequently in cells treated with either CAF-1-targeted siRNA. Arrows indicate examples of early, middle, and late replication sites (▹, early S-phase patterns; →, middle S-phase patterns; ▸, late S-phase patterns). (C) Distribution of S-phase cells displaying early, mid, or late S-phase replication patterns 48 h after transfection. Three hundred S-phase cells were counted per sample, and the results are the mean from two independent experiments.
Fig. 4.
Checkpoint activation in CAF-1-depleted cells. (A) Western analysis of total cell lysate after CAF-1 RNAi to monitor checkpoint activation by using phospho-specific antibodies to p53 S15, Chk1 S317, and Chk2 T68. Overall Chk2 levels do not change (data not shown). Extract of cells treated with hydroxyurea (20 mM) for 24 h, which leads to phosphorylation of all three proteins, was included as a positive control. (B) Time course of checkpoint activation and BrdUrd incorporation in control and CAF-1-depleted cells. Three coverslips were seeded per well and transfected according to the standard protocol. Cells were collected at 24, 36, 48, and 60 h after transfection, pulsed for 10 min with BrdUrd, and stained with phospho-specific antibodies against S15 of p53, S317 of Chk1, or BrdUrd. Cells in the 24-h time point had only been transfected once. Some loss in BrdUrd incorporation was seen by 36 h in comparison to control, and BrdUrd incorporation drops dramatically at 48 h after transfection. (C) Plot comparing checkpoint activation and BrdUrd incorporation in individual CAF-1 knockdown cells normalized to control transfected cells. Three hundred cells were counted per time point for each antibody for control and CAF-1 targeting oligo 1, and the results shown are the mean for two experiments. All cells displaying checkpoint activation above background and only brightly BrdUrd-labeled cells were scored as positive. Significant checkpoint activation was not seen until 48 h after transfection.
Fig. 5.
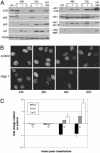
Replication coupled chromatin assembly activity depends on CAF-1. (A) Western blot analysis of CAF-1 in HeLa nuclear and S100 fractions after RNAi. (B) One-step chromatin assembly assays to monitor the ability of control and CAF-1-depleted nuclear extracts to assemble replicating SV40 DNA into nucleosomes. pSV011 DNA containing the SV40 origin was replicated in a replication mix containing 32P-dATP to label replicated DNA and was assembled into chromatin with nuclear extract or recombinant CAF-1. DNA packaged into chromatin accumulated negative supercoils and runs with enhanced mobility relative to unassembled DNA [compare lane 1 (low CAF-1) to lanes 2–4 (sufficient CAF-1)]. The bulk of unreplicated DNA visualized by ethidium bromide staining (Lower) remained relaxed, indicating that the CAF-1 preferentially loaded nucleosomes onto replicating DNA. When CAF-1 was depleted by RNAi (lanes 5–7 and 9–11), assembly did not occur. Addition of recombinant CAF-1 restored assembly activity in these extracts (lanes 8 and 12). Nuclear extract (10–30 μg) was titrated into each set of reactions. (C) Two-step chromatin assembly assays demonstrate that the S100 from CAF-1 knockdown cells contain all accessory factors (including histones H3 and H4) required to assemble chromatin. SV40 DNA was replicated in a S100 replication extract, purified away from proteins by gel filtration and incubated with S100 from control or p150-depleted extracts in the presence or absence of CAF-1. Twenty-five and 50 μg of extract were used in each reaction. Under these conditions, assembly into chromatin required CAF-1 (compare lanes 3 and 4, –/+ CAF-1) and S100 (compare lanes 2 and 4). Unreplicated DNA seen in the ethidium stain (Lower) was not assembled into chromatin and remained largely relaxed.
Similar articles
- Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis.
Nabatiyan A, Krude T. Nabatiyan A, et al. Mol Cell Biol. 2004 Apr;24(7):2853-62. doi: 10.1128/MCB.24.7.2853-2862.2004. Mol Cell Biol. 2004. PMID: 15024074 Free PMC article. - Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells.
Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T. Takami Y, et al. Mol Biol Cell. 2007 Jan;18(1):129-41. doi: 10.1091/mbc.e06-05-0426. Epub 2006 Oct 25. Mol Biol Cell. 2007. PMID: 17065558 Free PMC article. - An analysis of CAF-1-interacting proteins reveals dynamic and direct interactions with the KU complex and 14-3-3 proteins.
Hoek M, Myers MP, Stillman B. Hoek M, et al. J Biol Chem. 2011 Mar 25;286(12):10876-87. doi: 10.1074/jbc.M110.217075. Epub 2011 Jan 5. J Biol Chem. 2011. PMID: 21209461 Free PMC article. - Chromatin assembly during DNA replication in somatic cells.
Krude T. Krude T. Eur J Biochem. 1999 Jul;263(1):1-5. doi: 10.1046/j.1432-1327.1999.00508.x. Eur J Biochem. 1999. PMID: 10429179 Review. - Chromatin assembly during S phase: contributions from histone deposition, DNA replication and the cell division cycle.
Krude T, Keller C. Krude T, et al. Cell Mol Life Sci. 2001 May;58(5-6):665-72. doi: 10.1007/pl00000890. Cell Mol Life Sci. 2001. PMID: 11437228 Free PMC article. Review.
Cited by
- Cac1 WHD and PIP domains have distinct roles in replisome progression and genomic stability.
Tsirkas I, Dovrat D, Lei Y, Kalyva A, Lotysh D, Li Q, Aharoni A. Tsirkas I, et al. Curr Genet. 2021 Feb;67(1):129-139. doi: 10.1007/s00294-020-01113-8. Epub 2020 Oct 6. Curr Genet. 2021. PMID: 33025160 - Induction of CAF-1 expression in response to DNA strand breaks in quiescent human cells.
Nabatiyan A, Szüts D, Krude T. Nabatiyan A, et al. Mol Cell Biol. 2006 Mar;26(5):1839-49. doi: 10.1128/MCB.26.5.1839-1849.2006. Mol Cell Biol. 2006. PMID: 16479003 Free PMC article. - Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis.
Nabatiyan A, Krude T. Nabatiyan A, et al. Mol Cell Biol. 2004 Apr;24(7):2853-62. doi: 10.1128/MCB.24.7.2853-2862.2004. Mol Cell Biol. 2004. PMID: 15024074 Free PMC article. - Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C.
Franco AA, Lam WM, Burgers PM, Kaufman PD. Franco AA, et al. Genes Dev. 2005 Jun 1;19(11):1365-75. doi: 10.1101/gad.1305005. Epub 2005 May 18. Genes Dev. 2005. PMID: 15901673 Free PMC article. - Yeast chromatin assembly complex 1 protein excludes nonacetylatable forms of histone H4 from chromatin and the nucleus.
Glowczewski L, Waterborg JH, Berman JG. Glowczewski L, et al. Mol Cell Biol. 2004 Dec;24(23):10180-92. doi: 10.1128/MCB.24.23.10180-10192.2004. Mol Cell Biol. 2004. PMID: 15542829 Free PMC article.
References
- Jackson, V. (1990) Biochemistry 29, 719–731. - PubMed
- Gasser, R., Koller, T. & Sogo, J. M. (1996) J. Mol. Biol. 258, 224–239. - PubMed
- Smith, S. & Stillman, B. (1989) Cell 58, 15–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous